Taxonomic group
advertisement
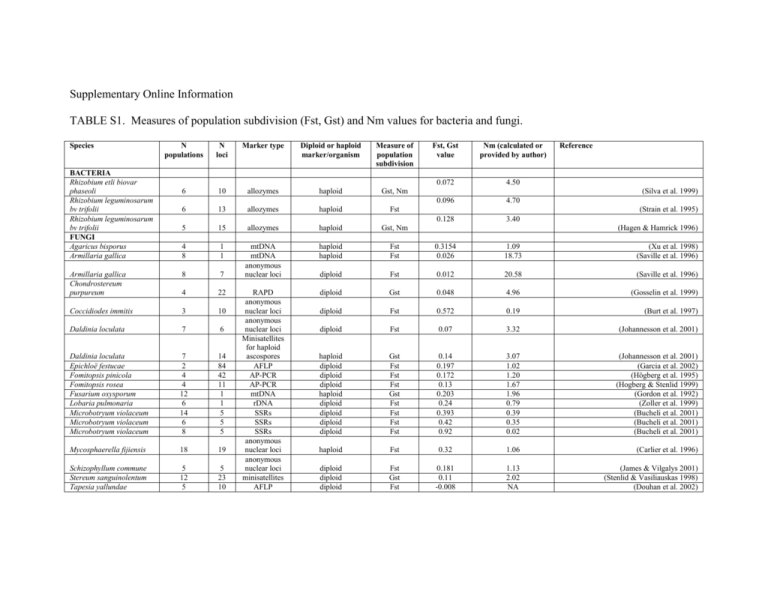
Supplementary Online Information TABLE S1. Measures of population subdivision (Fst, Gst) and Nm values for bacteria and fungi. Species BACTERIA Rhizobium etli biovar phaseoli Rhizobium leguminosarum bv trifolii Rhizobium leguminosarum bv trifolii FUNGI Agaricus bisporus Armillaria gallica N populations N loci Marker type Diploid or haploid marker/organism Measure of population subdivision 6 10 allozymes haploid Gst, Nm 6 13 allozymes haploid Fst, Gst value Nm (calculated or provided by author) 0.072 4.50 0.096 4.70 0.128 3.40 Reference (Silva et al. 1999) Fst (Strain et al. 1995) 5 15 allozymes haploid Gst, Nm (Hagen & Hamrick 1996) 4 8 1 1 haploid haploid Fst Fst 0.3154 0.026 1.09 18.73 (Xu et al. 1998) (Saville et al. 1996) Armillaria gallica Chondrostereum purpureum 8 7 mtDNA mtDNA anonymous nuclear loci diploid Fst 0.012 20.58 (Saville et al. 1996) 4 22 diploid Gst 0.048 4.96 (Gosselin et al. 1999) Coccidiodes immitis 3 10 diploid Fst 0.572 0.19 (Burt et al. 1997) Daldinia loculata 7 6 diploid Fst 0.07 3.32 (Johannesson et al. 2001) Daldinia loculata Epichloё festucae Fomitopsis pinicola Fomitopsis rosea Fusarium oxysporum Lobaria pulmonaria Microbotryum violaceum Microbotryum violaceum Microbotryum violaceum 7 2 4 4 12 6 14 6 8 14 84 42 11 1 1 5 5 5 haploid diploid diploid diploid haploid diploid diploid diploid diploid Gst Fst Fst Fst Gst Fst Fst Fst Fst 0.14 0.197 0.172 0.13 0.203 0.24 0.393 0.42 0.92 3.07 1.02 1.20 1.67 1.96 0.79 0.39 0.35 0.02 (Johannesson et al. 2001) (Garcia et al. 2002) (Högberg et al. 1995) (Hogberg & Stenlid 1999) (Gordon et al. 1992) (Zoller et al. 1999) (Bucheli et al. 2001) (Bucheli et al. 2001) (Bucheli et al. 2001) Mycosphaerella fijiensis 18 19 haploid Fst 0.32 1.06 (Carlier et al. 1996) Schizophyllum commune Stereum sanguinolentum Tapesia yallundae 5 12 5 5 23 10 RAPD anonymous nuclear loci anonymous nuclear loci Minisatellites for haploid ascospores AFLP AP-PCR AP-PCR mtDNA rDNA SSRs SSRs SSRs anonymous nuclear loci anonymous nuclear loci minisatellites AFLP diploid diploid diploid Fst Gst Fst 0.181 0.11 -0.008 1.13 2.02 NA (James & Vilgalys 2001) (Stenlid & Vasiliauskas 1998) (Douhan et al. 2002) 2 TABLE S2. Measures of population subdivision (Fst, Gst, Φst) and Nm values for plants. Fst, Gst, Φst value Nm (calculated or provided by author) Mating system Distribution Range covered Measure of population subdivision outcrossing regional species-wide Gst 0.133 3.26 both unknown regional regional Φst 0.6372 0.14 Reference (Vendramin et al. 1999) (Liao & Hsiao 1998) both outcrossing regional regional Fst, Nm 0.107 2.08 (Ge & Sun 1999) SSRs both mixed regional regional Gst, Nm 0.178 1.16 (Ge & Sun 1999) 45 RAPDs both outcrossing regional local Φst 0.325 0.52 3 138 RAPDs both endemic regional Fst 0.2935 0.60 Anisantha sterilis 3 9 SSRs both outcrossing selfing and/or clonal (Wolff et al. 1997) (Kwon & Morden 2002) widespread local Fst 0.3108 0.55 (Green et al. 2001) Anthyllis montana 16 1199 AFLP both outcrossing regional species-wide Fst 0.4237 0.34 Antirrhinum charidemi 5 11 allozymes both outcrossing endemic local Gst 0.054 4.38 Antirrhinum valentinum Arceuthobium americanum 5 11 allozymes both outcrossing endemic species-wide Gst 0.48 0.27 51 85 AFLP both outcrossing regional species-wide Gst 0.303 0.58 Argania spinosa 19 1 cpDNA seed outcrossing endemic species-wide Gst 0.6 0.33 (Kropf et al. 2002) (Mateu-Andres & Segarra-Moragues 2000) (Mateu-Andres & Segarra-Moragues 2000) (Jerome & Ford 2002) (ElMousadik & Petit 1996) Argyroxiphium kauense 3 7 SSRs both outcrossing endemic species-wide Fst 0.141 1.52 Aster tripolium Astragalus cremnophylax 8 33 RAPDs both mixed regional regional Φst 0.175 1.18 (Friar et al. 2001) (Kruger et al. 2002) 3 220 AFLP both outcrossing endemic species-wide Fst 0.41 0.36 (Travis et al. 1996) Avicennia germinans 14 349 AFLP both outcrossing widespread species-wide Φst, Nm 0.41 0.41 Avicennia marina 14 3 SSRs both unknown widespread species-wide Fst 0.41 0.36 Beta vulgaris 40 1 cpDNA seed mixed regional regional Fst, Nm 0.43 1.33 Buchloё dactyloides 4 10 allozymes both mixed regional regional Φst 0.547 0.21 (Dodd et al. 2002) (Maguire et al. 2000) (Forcioli et al. 1998) (Peakall et al. 1995) Buchloё dactyloides 4 98 RAPDs both mixed regional regional Φst 0.681 0.12 (Peakall et al. N pop’ns N loci Marker Pollen or seed? Abies alba 17 1 cpSSRs pollen Acorus gramineus 12 17 RAPDs Aegiceras corniculatum 10 9 allozymes Aegiceras corniculatum 10 18 Alkanna orientalis 4 Alphitonia ponderosa Species 3 1995) Caesalpinia echinata 5 134 RAPDs both outcrossing regional regional Φst 0.58 0.18 Calluna vulgaris Calycophyllum spruceanum 23 1 cpDNA seed mixed regional species-wide Gst 0.473 0.56 9 65 AFLP both outcrossing regional regional Fst 0.118 1.87 Carapa guianensis 3 3 SSRs both outcrossing regional local Rst, Nm 0.017 14.10 Carex bigelowii 2 10 allozymes both regional regional Gst 0.13 1.67 Carex ensifolia 8 10 allozymes both mixed selfing and/or clonal regional regional Gst 0.151 1.41 Carex lugens 3 10 allozymes both mixed regional regional Gst 0.057 4.14 Carex stans 6 10 allozymes both mixed regional regional Gst 0.425 0.34 Carpentaria acuminata 37 14 allozymes both mixed endemic species-wide Fst, Nm 0.379 0.39 Caryocar brasiliense 10 10 SSRs both outcrossing regional species-wide Fst 0.07 3.32 Castanea sativa 38 1 cpDNA seed outcrossing widespread species-wide Gst 0.43 0.66 Cedrela odorata 10 97 RAPDs both outcrossing regional regional Φst 0.349 0.47 Centaurea corymbosa 6 5 allozymes both outcrossing endemic species-wide Fst 0.35 0.46 Centaurea corymbosa 6 6 SSRs both outcrossing endemic species-wide Fst 0.23 0.84 Ceramium tenuicorne Cerastium fischerianum fischerianum Cerastium fischerianum molle Cladophoropsis membranacea 9 42 RAPDs both mixed regional regional Fst 0.31 0.56 9 110 RAPDs both widespread local Φst 0.3909 0.39 8 71 RAPDs both endemic local Φst 0.3493 0.47 6 8 SSRs both mixed selfing and/or clonal selfing and/or clonal widespread local Fst 0.205 0.97 Colubrina oppositifolia 4 155 RAPDs both outcrossing endemic regional Fst 0.4573 0.30 Corylus avellana 26 1 cpSSRs seed outcrossing regional species-wide Fst 0.857 0.08 Corylus avellana 26 1 cpRFLP seed outcrossing regional species-wide Fst 0.08 5.75 Cycas taitungensis 2 1 cpDNA seed outcrossing endemic species-wide Fst, Nm 0.0056 90.41 (Cardoso et al. 1998) (Rendell & Ennos 2002) (Russell et al. 1999) (Dayanandan et al. 1999) (Stenstrom et al. 2001) (Stenstrom et al. 2001) (Stenstrom et al. 2001) (Stenstrom et al. 2001) (Shapcott 1998) (Collevatti et al. 2001) (Fineschi et al. 2000) (Gillies et al. 1997) (Freville et al. 2001) (Freville et al. 2001) (Gabrielsen et al. 2002) (Maki & Horie 1999) (Maki & Horie 1999) (Van der Strate et al. 2002) (Kwon & Morden 2002) (Palme & Vendramin 2002) (Palme & Vendramin 2002) (Huang et al. 2001) 4 Cycas taitungensis 2 1 mtDNA seed outcrossing endemic species-wide Fst, Nm 0.021 23.18 Cyclobalanopsis glauca 25 1 cpDNA seed outcrossing regional regional Gst 0.612 0.32 Cypripedium calceolus 3 4 allozymes both mixed endemic local Fst, Nm 0.014 18.00 Dendropanax arboreus 5 29 RAPDs both outcrossing widespread local Φst 0.032 7.56 Dicorynia guianensis 12 1 cpDNA seed outcrossing endemic regional Nst 0.25 1.50 Dryas integrifolia 42 1 cpDNA seed unknown widespread species-wide Gst 0.44 0.64 Elliottia racemosa 10 9 allozymes both mixed endemic species-wide Gst, Nm 0.18 1.20 Eritrichium nanum 20 884 AFLP both outcrossing regional species-wide Fst 0.166 1.26 Erodium paularense 3 26 RAPDs both outcrossing endemic species-wide Φst 0.1974 1.02 Erodium paularense 2 127 RAPDs both outcrossing endemic biregional Φst 0.2683 0.68 Eryngium alpinum 14 62 AFLP both mixed regional local Fst 0.42 0.35 Euterpe edulis 11 429 AFLP both mixed regional species-wide Fst 0.426 0.34 Fagus sylvatica 6 13 allozymes both outcrossing regional local Gst 0.0091 27.22 Festuca pratensis 6 36 RAPDs both unknown widespread regional Φst 0.2603 0.71 Fitzroya cupressoides 12 54 RAPDs both outcrossing endemic species-wide Φst 0.1438 1.49 Fraxinus excelsior 10 6 SSRs both outcrossing regional regional Fst 0.087 2.62 Fraxinus excelsior 12 5 SSRs both outcrossing regional regional Fst 0.043 5.56 Grevillea macleayana 6 6 SSRs both mixed endemic regional Fst 0.218 0.90 Grevillea scapigera 5 167 RAPDs both outcrossing endemic species-wide Φst 0.1305 1.67 Gymnadenia conopsea Haplostachys halplostachya 10 3 SSRs both outcrossing regional local Fst, Nm 0.06 3.90 3 54 RAPDs both endemic local Gst 0.079 2.91 Heteropogon contortus 6 33 RAPDs both outcrossing selfing and/or clonal widespread regional Φst 0.3185 0.53 Hippophae rhamnoides 11 156 RAPDs both outcrossing regional regional Φst 0.209 0.95 (Huang et al. 2001) (Huang et al. 2002) (Brzosko et al. 2002) (Schierenbeck et al. 1997) (Caron et al. 2000) (Tremblay & Schoen 1999) (Godt & Hamrick 1999) (Stehlik et al. 2001) (Martin et al. 1997) (Martin et al. 1999) (Gaudeul et al. 2000) (Cardoso et al. 2000) (Sander et al. 2000) (Kolliker et al. 1998) (Allnutt et al. 1999) (Heuertz et al. 2001) (Morand et al. 2002) (England et al. 2002) (Rossetto et al. 1995) (Gustafsson 2000) (Morden & Loeffler 1999) (Carino & Daehler 1999) (Bartish et al. 1999) 5 (Dawson et al. 1993) (Turpeinen et al. 2001) (Chiang & Schaal 1999) (Schierenbeck et al. 1997) (Arafeh et al. 2002) Hordeum spontaneum 20 7 RAPDs both outcrossing regional regional Gst 0.67 0.12 Hordeum spontaneum 10 18 SSRs both outcrossing regional regional Gst 0.36 0.44 Hylocomium splendens 3 1 nrDNA both mixed widespread regional Fst 0.296 0.59 Inga thibaudiana 3 29 RAPDs both outcrossing regional local Φst 0.009 27.53 Iris haynei/atrofusca 9 132 RAPDs both outcrossing endemic species-wide Gst, Nm 0.18 1.14 Isotoma petraea 6 60 RAPDs both mixed regional regional Gst 0.04 Kandelia candel 4 1 cpDNA seed outcrossing regional species-wide Fst, Nm 36.20 Kandelia candel 4 1 mtDNA seed outcrossing regional species-wide Fst, Nm 0.875 0.0232 5 0.0187 5 Lambertia orbifolia 2 1 cpDNA seed mixed regional biregional Gst 0.855 0.08 Leucopogon obtectus 4 82 RAPDs both outcrossing endemic species-wide Fst 0.133 1.63 Leucopogon obtectus 4 264 AFLP both outcrossing endemic species-wide Fst 0.103 2.18 Limonium dufourii 6 33 RAPDs both endemic species-wide Φst 0.656 0.13 Limonium dufourii 6 51 AFLP both selfing and/or clonal selfing and/or clonal endemic species-wide Fst 0.243 0.78 16? 43 RAPDs both mixed regional local Φst, Nm 0.1864 1.10 Medicago sativa 7 1 mtDNA seed regional regional Fst 0.12 3.67 Medicago truncatula 3 5 SSRs both outcrossing selfing and/or clonal regional local Fst 0.37 0.43 Melaleuca alternifolia 40 5 SSRs both outcrossing regional species-wide Fst, Nm 0.072 3.40 Metrosideros bartlettii 3 286 AFLP both mixed endemic local Fst 0.144 1.49 Oryza glumaepatula 37 12 allozymes both outcrossing regional regional Fst 0.346 0.47 (Byrne et al. 1999) (Zawko et al. 2001) (Zawko et al. 2001) (Palacios & GonzalezCandelas 1997) (Palacios et al. 1999) (Grashof-Bokdam et al. 1998) (Muller et al. 2001) (Bonnin et al. 2001) (Rossetto et al. 1999) (Drummond et al. 2000) (Akimoto et al. 1998) Oryza glumaepatula 4 156 RAPDs both outcrossing regional regional Fst 0.67 0.12 (Buso et al. 1998) Oryza glumaepatula 4 3 allozymes both outcrossing regional regional Fst 0.31 0.56 Parnassia palustris 14 6 allozymes both outcrossing widespread regional Fst 0.16 1.31 Parnassia palustris 14 1 cpDNA seed outcrossing widespread regional Fst 0.79 0.13 (Buso et al. 1998) (Bonnin et al. 2002) (Bonnin et al. 2002) Phyteuma 69 240 AFLP both outcrossing regional species-wide Fst 0.59 0.17 (Schönswetter et Lonicera periclymenum 28.60 (Bussell 1999) (Chiang et al. 2001) (Chiang et al. 2001) 6 globulariifolium al. 2002) Picea abies 8 7 SCAR both outcrossing widespread regional Fst 0.118 1.87 Picea abies 95 3 SSRs both outcrossing widespread species-wide Fst 0.1451 1.47 Picea abies 36 1 mtDNA seed outcrossing widespread regional Fst 0.41 0.72 Picea glauca 6 13 allozymes both outcrossing widespread regional Fst 0.014 17.61 Picea glauca 6 11 ESTP both outcrossing widespread regional Fst 0.02 12.25 (Scotti et al. 2000) (Bucci & Vendramin 2000) (Gugerli et al. 2001) (Jaramillo-Correa et al. 2001) (Jaramillo-Correa et al. 2001) Pinanga aristata 7 8 allozymes both mixed endemic local Fst, Nm 0.21 0.93 (Shapcott 1999) Pinanga dumetosa Pinanga sp. aff. brevipes 7 9 allozymes both mixed endemic local Fst, Nm 0.099 2.27 (Shapcott 1999) 7 8 allozymes both mixed endemic local Fst, Nm 0.261 0.70 (Shapcott 1999) Pinanga tenella 7 9 allozymes both mixed endemic local Fst, Nm 0.148 1.44 (Shapcott 1999) Pinanga veitchii 7 8 allozymes both mixed endemic local Fst, Nm 1.03 Pinus albicaulis 41 1 cpSSRs pollen outcrossing regional species-wide Φst 0.194 0.0463 6 10.29 (Shapcott 1999) (Richardson et al. 2002) Pinus brutia 8 1 cpSSRs pollen outcrossing regional regional Gst 0.289 1.23 (Bucci et al. 1998) Pinus eldarica 2 1 cpSSRs pollen outcrossing regional regional Gst 0.068 6.85 Pinus flexilis 40 1 mtDNA seed outcrossing regional species-wide Fst, Nm 0.8 0.12 (Bucci et al. 1998) (Mitton et al. 2000) Pinus halepensis 10 1 cpSSRs pollen outcrossing regional regional Gst 0.308 1.12 (Bucci et al. 1998) Pinus oocarpa 10 131 RAPDs both outcrossing regional regional Fst 0.122 1.80 (Diaz et al. 2001) Pinus oocarpa 10 392 AFLP both outcrossing regional regional Fst 0.086 2.66 Pinus pinaster 12 19 AFLP both outcrossing regional regional Gst 0.047 5.07 Pinus pinaster 12 1 cpSSRs pollen outcrossing regional regional Gst 0.039 12.32 (Diaz et al. 2001) (Ribeiro et al. 2002) (Ribeiro et al. 2002) Pinus resinosa 7 1 cpSSRs pollen outcrossing regional species-wide Gst 0.121 3.63 Pinus resinosa 10 10 cpDNA pollen outcrossing regional regional Fst 0.56 0.39 Pinus sylvestris 20 1 mtDNA seed outcrossing widespread regional Fst 0.37 0.85 Pinus sylvestris 7 1 mtDNA seed outcrossing widespread regional Fst 0.817 0.11 Pinus sylvestris 23 1 seed regional Φst 0.59 0.35 10 73 both outcrossing selfing and/or clonal widespread Plantago intermedia mtDNA RAPDs and inter-SSRs widespread species-wide Φst 0.672 0.12 (Echt et al. 1998) (Walter & Epperson 2001) (Sinclair et al. 1999) (Sinclair et al. 1999) (Soranzo et al. 2000) (Morgan-Richards & Wolff 1999) Plantago intermedia 9 8 allozymes both selfing and/or widespread species-wide Fst 0.777 0.07 (Morgan-Richards 7 clonal Plantago major 9 73 RAPDs and inter-SSRs Plantago major 9 8 allozymes both selfing and/or clonal selfing and/or clonal Plathymenia reticulata 6 51 RAPDs both Populus tremuloides 23 132 RAPDs Posidonia oceanica 17 6 Potamogeton pectinatus 40 Protium glabrum & Wolff 1999) widespread species-wide Φst 0.397 0.38 widespread species-wide Fst 0.232 0.83 unknown regional regional Φst, Nm 0.12 2.34 both outcrossing widespread local Fst 0.34 0.49 SSRs both endemic regional Fst, Nm 0.104 2.15 62 ISSR both mixed selfing and/or clonal regional regional Φst 0.496 0.25 3 20 RAPDs both outcrossing regional local Φst -0.003 -83.58 Prunus africana 10 48 RAPDs both outcrossing regional species-wide Φst 0.734 0.09 Prunus mahaleb 7 73 RAPDs both mixed regional local Gst, Nm 0.1935 1.19 Pseudotsuga menziesii 3 16 RAPDs both outcrossing regional regional Gst 0.73 0.09 Pseudotsuga menziesii 3 19 allozymes both outcrossing regional regional Gst 0.26 0.71 Pseudotsuga menziesii 3 1 mtDNA seed outcrossing regional regional Gst 0.62 0.31 Pseudotsuga menziesii 3 1 seed outcrossing regional regional Gst 0.72 0.19 Pseudotsuga menziesii 3 1 mtDNA nuclear RAPDs both outcrossing regional regional Gst 0.25 0.75 Quercus gambelii 3 127 minisatellites both mixed regional local Fst 0.243 0.78 Quercus ilex 24 1 cpDNA seed outcrossing regional regional Gst 0.33 1.02 Quercus petraea 21 8 allozymes both outcrossing regional species-wide Gst 0.027 9.01 Quercus petraea 21 31 RAPDs both outcrossing regional species-wide Gst 0.024 10.17 Quercus suber Rhododendron championiae 73 1 cpDNA seed outcrossing regional regional Gst 0.84 0.10 3 18 allozymes both outcrossing regional local Fst, Nm 0.198 1.01 Rhododendron farrerae Rhododendron honkongense 5 15 allozymes both outcrossing widespread local Fst, Nm 0.128 1.70 3 13 allozymes both outcrossing regional local Fst, Nm 0.191 1.06 both (Morgan-Richards & Wolff 1999) (Morgan-Richards & Wolff 1999) (Lacerda et al. 2001) (Stevens et al. 1999) (Procaccini et al. 2001) (King et al. 2002) (Schierenbeck et al. 1997) (Dawson & Powell 1999) (Jordano & Godoy 2000) (Aagaard et al. 1995) (Aagaard et al. 1995) (Aagaard et al. 1995) (Aagaard et al. 1998) (Aagaard et al. 1998) (Kumar & Rogstad 1998) (Belahbib et al. 2001) (LeCorre et al. 1997) (LeCorre et al. 1997) (Belahbib et al. 2001) (Ng & Corlett 2000) (Ng & Corlett 2000) (Ng & Corlett 2000) 8 Rhododendron moulmainense Rhododendron simiarum 4 15 allozymes both outcrossing regional local Fst, Nm 0.081 2.83 2 16 allozymes both outcrossing regional local Fst, Nm 0.393 0.39 Rhododendron simsii 8 15 allozymes both outcrossing widespread local Fst, Nm 0.056 4.21 Salicornia ramosissima 6 72 RAPDs both regional regional Φst 0.35 0.46 Sarconeurum glaciale 4 83 RAPDs both regional local Φst 0.273 0.67 Saxifraga cernua 2 38 RAPDs both widespread local Φst 0.4087 0.36 Saxifraga cespitosa 30 37 RAPDs both mixed selfing and/or clonal selfing and/or clonal selfing and/or clonal regional regional Φst 0.71 0.10 Saxifraga oppositifolia 18 35 RAPDs both outcrossing widespread regional Φst 0.36 0.44 Saxifraga oppositifolia 10 84 RAPDs both outcrossing widespread local Φst 0.05 4.75 Saxifraga oppositifolia 15 1 cpDNA seed outcrossing widespread regional Fst 0.213 1.85 Saxifraga oppositifolia Schizachyrium scoparium 15 151 RAPDs both outcrossing widespread regional Fst 0.152 1.39 (Ng & Corlett 2000) (Ng & Corlett 2000) (Ng & Corlett 2000) (Kruger et al. 2002) (Skotnicki et al. 1999) (Gabrielsen & Brochmann 1998) (Tollefsrud et al. 1998) (Gabrielsen et al. 1997) (Gugerli et al. 1999) (Holderegger et al. 2002) (Holderegger et al. 2002) 4 60 RAPDs both regional regional Fst 0.05 4.75 (Huff et al. 1998) Scutellaria indica 20 8 allozymes both regional regional Fst 0.937 0.02 (Sun 1999) Scutellaria indica 10 ? RAPDs both unknown selfing and/or clonal selfing and/or clonal regional regional Gst 0.81 0.06 Senecio gallicus 11 9 RAPDs both regional regional Fst 0.339 0.49 Senecio vulgaris 9 26 RAPDs both widespread local Φst, Nm 0.382 0.40 Senecio vulgaris 10 183 AFLP both outcrossing selfing and/or clonal selfing and/or clonal widespread local Φst 0.49 0.26 Silene acaulis 10 7 allozymes both outcrossing widespread regional Fst, Nm 0.141 1.52 Silene paradoxa 8 59 RAPDs both outcrossing regional local Φct 0.116 1.91 Silene paradoxa 8 1 cpSSRs seed outcrossing regional local Φst 0.528 0.45 (Sun 1999) (Comes & Abbott 2000) (Muller-Scharer & Fischer 2001) (Steinger et al. 2002) (Abbott et al. 1995) (Mengoni et al. 2000) (Mengoni et al. 2001) Sorbus aucuparia 6 1 cpDNA seed outcrossing widespread regional Gst 0.29 1.22 Sticherus flabellatus 8 469 AFLP both unknown regional regional Fst 0.783 0.07 Swietenia humilis 2 10 SSRs both outcrossing regional local Fst 0.036 6.69 (Raspe et al. 2000) (Keiper & McConchie 2000) (White & Powell 1997) 9 Swietenia humilis 4 10 SSRs both outcrossing regional local Rst, Nm 0.032 8.90 Symphonia globulifera 2 3 SSRs both mixed regional local Fst 0.031 7.81 Symplocos laurina 2 380 SSRs both unknown endemic biregional Gst, Nm 0.463 0.45 Trollius europaeus 18 117 AFLP both outcrossing regional species-wide Fst 0.39 0.39 Viola riviniana 6 45 RAPDs both mixed regional local Φst 0.41 0.36 Vouacapoua americana 14 1 cpDNA seed outcrossing regional local Gst 0.89 0.06 Vouacapoua americana 4 8 SSRs both outcrossing regional local Fst 0.11 2.02 Vouacapoua americana 3 8 SSRs both regional local Fst 0.04 6.00 Wyethia reticulata 4 21 RAPDs both endemic species-wide Fst, Nm 0.25 0.80 Wyethia reticulata Yushania niitakayamensis 4 9 allozymes both endemic species-wide Fst, Nm 0.19 1.10 13 43 RAPDs both outcrossing selfing and/or clonal selfing and/or clonal selfing and/or clonal regional regional Φst 0.2007 1.00 Zostera marina 12 6 SSRs both mixed widespread species-wide Φst 0.29 0.61 (White et al. 1999) (Aldrich et al. 1998) (Deshpande et al. 2001) (Despres et al. 2002) (Auge et al. 2001) (Dutech et al. 2000) (Dutech et al. 2002) (Dutech et al. 2002) (Ayres & Ryan 1997) (Ayres & Ryan 1997) (Hsiao & Lee 1999) (Reusch et al. 2000) 10 TABLE S3. Measures of population subdivision (Fst, Gst, Φst) and Nm values for animals. Fst,Gst,Φst value species-wide species-wide regional Measure of population subdivision Fst Φst Fst 0.724 0.023 0.356 Nm (calculated or provided by author) 0.19 21.24 0.90 endemic regional endemic widespread widespread widespread species-wide species-wide regional local local local Fst Fst Fst Fst, Nm Fst, Nm Fst 0.809 0.332 0.308 0.033 0.063 0.056 0.12 1.01 1.12 7.20 3.70 4.21 outcrossing regional species-wide Kst 0.247 1.52 outcrossing outcrossing outcrossing outcrossing regional endemic endemic endemic regional species-wide species-wide species-wide Gst Fst Fst Fst 0.03 0.68 0.103 0.185 8.08 0.24 2.18 1.10 6 1 1 allozymes mtDNA SSRs minisatellites SSRs + minisatellites mtDNA mtDNA outcrossing outcrossing outcrossing endemic endemic regional species-wide species-wide local Fst Fst Fst, Nm 0.119 0.639 0.294 1.85 0.28 1.20 4 2 3 4 4 5 10 11 11 11 1 5 11 SSRs SSRs SSRs SSRs mtDNA SSRs SSRs outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing widespread regional endemic regional widespread widespread widespread regional local local local local local regional Fst Fst Fst Fst Fst Fst Fst 0.106 0.007 0.188 0.052 0.066 0.013 0.1647 2.11 35.46 1.08 4.56 7.08 18.98 1.27 Aphis gossypii Aphodius fossor Apis mellifera Apis mellifera Arctocephalus gazella Arctocephalus tropicalis 18 14 5 5 8 8 37 7 8 1 1 1 RAPDs allozymes SSRs mtDNA mtDNA mtDNA mixed outcrossing mixed mixed outcrossing outcrossing widespread widespread widespread widespread regional regional regional regional local local species-wide species-wide Φst Fst Fst Fst Φst Φst 0.879 0.011 0.0772 0.1907 0.074 0.19 0.03 22.48 2.99 2.12 6.26 2.13 Atypus affinis Austropotamobius 4 25 15 19 allozymes allozymes outcrossing outcrossing endemic regional species-wide regional Fst, Nm Fst 0.052 0.925 4.60 0.02 Species N pop’ns N loci Marker Mating system Distribution Range covered Acanthurus bahianus Acanthurus chirurgus Acanthurus coeruleus 6 3 4 1 1 1 mtDNA mtDNA mtDNA outcrossing outcrossing outcrossing regional regional regional Achatinella mustelina Acipenser brevirostrum Acrossocheilus paradoxus Aedes aegypti Aedes aegypti Aedes aegypti 18 9 12 3 3 20 1 1 1 137 6 6 mtDNA mtDNA mtDNA AFLP RFLP SSRs unknown outcrossing outcrossing outcrossing outcrossing outcrossing Aepyceros melampus 12 1 mtDNA Aglais urticae Amblyrhynchus cristatus Amblyrhynchus cristatus Amblyrhynchus cristatus 9 22 22 6 5 1 3 3 Amblyrhynchus cristatus Andrias davidianus Anolis marmoratus 6 6 5 Anopheles arabiensis Anopheles dirus A Anopheles dirus C Anopheles dirus D Anopheles gambiae Anopheles gambiae Anopheles gambiae Reference (Rocha et al. 2002) (Rocha et al. 2002) (Rocha et al. 2002) (Holland & Hadfield 2002) (Grunwald et al. 2002) (Wang et al. 2000) (Yan et al. 1999) (Yan et al. 1999) (Huber et al. 2002) (Nersting & Arctander 2001) (Vandewoestijne et al. 1999) (Rassmann et al. 1997) (Rassmann et al. 1997) (Rassmann et al. 1997) (Rassmann et al. 1997) (Murphy et al. 2000) (Schneider 1996) (Onyabe & Conn 2001) (Walton et al. 2001) (Walton et al. 2001) (Walton et al. 2001) (Lehmann et al. 1997) (Lehmann et al. 1997) (Pinto et al. 2002) (Vanlerberghe-Masuti & Chavigny 1998) (Roslin 2001) (De la Rua et al. 2001) (De la Rua et al. 2001) (Wynen et al. 2000) (Wynen et al. 2000) (Pedersen & Loeschcke 2001) (Largiader et al. 2000) 11 pallipes Axoclinus nigricaudus Batillariella estuarina Bemisia tabaci, biotype B Bemisia tabaci, biotype Q Bettongia tropica Bettongia tropica Biomphalaria glabrata 9 16 6 6 4 4 20 1 11 234 234 1 7 7 mtDNA allozymes RAPDs RAPDs mtDNA SSRs SSRs Biomphalaria pfeifferi 30 8 Birgus latro 8 Bombus pascuorum Bombus terrestris Brachyramphus marmoratus Bufo calamita Bufo canorus Calidris alpina Calidris melanotos Calidris pusilla Camponotus floridanus Canis lupus (Riginos & Nachman 2001) (Pudovskis et al. 2001) (Moya et al. 2001) (Moya et al. 2001) (Pope et al. 2000) (Pope et al. 2000) (Mavarez et al. 2002) (Charbonnel et al. 2002) endemic regional widespread widespread endemic endemic regional species-wide regional regional regional local local local Fst Fst Fst Fst Fst Fst Fst 0.536 0.455 0.325 0.273 0.561 0.062 0.23 0.43 0.30 0.52 0.67 0.39 3.78 0.84 SSRs outcrossing unknown outcrossing outcrossing outcrossing outcrossing mixed selfing and/or clonal regional Fst 0.8 0.06 1 mtDNA outcrossing widespread regional almost species-wide Gst, Nm 0.37 0.85 18 14 6 8 SSRs SSRs outcrossing outcrossing regional regional regional species-wide Fst Fst 0.0451 0.106 5.29 2.11 (Lavery et al. 1996) (Widmer & SchmidHempel 1999) (Estoup et al. 1996) 9 40 28 3 2 5 nuclear introns SSRs mtDNA RAPDs RAPDs RAPDs outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional regional endemic widespared regional regional regional local species-wide regional regional regional Fst Fst Φst Fst, Nm Fst, Nm Fst, Nm 0.093 0.365 0.76 0.26 0.27 0.16 2.44 0.43 0.16 0.71 0.27 1.31 (Friesen et al. 1997) (Rowe et al. 1998) (Shaffer et al. 2000) (Haig et al. 1997) (Haig et al. 1997) (Haig et al. 1997) 3 30 4 8 1 6 10 8 2835 1 DNA Fing. mtDNA mixed outcrossing widespread widespread regional species-wide Fst Fst 0.19 0.744 1.07 0.17 Canis lupus Canis simensis Capra ibex Capreolus capreolus Caretta caretta Ceratitis capitata 9 2 7 17 6 16 9 9 18 1 1 1 outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing widespread endemic regional widespread widespread widespread regional regional species-wide regional regional species-wide Fst Fst Fst Φst Φst Fst 0.091 0.057 0.167 0.63 0.331 0.296 2.50 4.14 1.25 0.29 1.01 0.59 Ceratitis capitata Cervus nippon Cervus nippon Chalcides viridanus Charadrius semipalmatus Chelonia mydas 20 3 14 17 3 9 1 1 9 2 9 1 SSRs SSRs SSRs mtDNA mtDNA Adh1 intron glucose-6phosphate dehydrogenase intron mtDNA SSRs mtDNA RAPDs mtDNA (Gadau et al. 1996) (Vilá et al. 1999) (Carmichael et al. 2001) (Gottelli et al. 1994) (Maudet et al. 2002) (Vernesi et al. 2002) (Laurent et al. 1998) (Gomulski et al. 1998) outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing widespread regional regional endemic regional widespread regional local regional species-wide regional regional Φst Φst Fst Nst Fst, Nm Nm 0.2 0.36 0.415 0.768 0.25 NA 1.00 0.89 0.35 0.15 0.75 0.64 Chioglossa lusitanica Chrysochus cobaltinus 17 5 7 1 allozymes mtDNA outcrossing outcrossing endemic regional species-wide regional Fst Φst 0.678 0.766 0.12 0.15 (He & Haymer 1999) (Nagata et al. 1998) (Goodman et al. 2001) (Brown et al. 2000) (Haig et al. 1997) (Encalada et al. 1996) (Alexandrino et al. 2000) (Dobler & Farrell 12 Clethrionomys glareolus Clevelandia ios 31 10 1 1 mtDNA mtDNA outcrossing outcrossing regional regional local regional Fst Φst 0.234 0.0281 1.64 17.29 Coelopa frigida 2 1 mtDNA outcrossing regional biregional Fst 0.76 0.16 Columbia inornata Coregonus lavaretus/oxyrhynchus Coregonus lavaretus/oxyrhynchus Corophium volutator 2 1 mtDNA outcrossing endemic local Gst, Nm 0.0956 1.18 1999) (Stacy et al. 1997) (Dawson et al. 2002) (MacDonald & Brookfield 2002) (Young & Allard 1997) 12 1 mtDNA outcrossing regional regional Fst 0.434 0.65 (Hansen et al. 1999) 12 6 6 32 SSRs RAPDs outcrossing unknown regional widespread regional local Fst Φst 0.106 0.2051 2.11 0.97 Corvus kubaryi 2 9 SSRs outcrossing endemic biregional Fst 0.281 0.64 Cottus gobio 12 31 allozymes outcrossing regional local Fst, Nm 0.35 0.56 Cottus gobio Cottus gobio Craterocephalus stercusmuscarum Crepidula atrasolea Crepidula convexa southern Crepidula convexanorthern Crepidula depressa Crepidula fornicata Crocidura russula Cucumaria miniata Cucumaria pseudocurata Culex pipiens Culex quinquefasciatus Culicoides bolitinos Culicoides imicola (ss) Culicoides loxodontis 69 44 9 10 allozymes SSRs outcrossing outcrossing regional regional regional species-wide Fst Fst 0.54 0.49 0.21 0.26 16 3 7 1 allozymes mtDNA outcrossing outcrossing regional regional regional regional Fst Fst 0.79 0.543 0.07 0.42 (Hansen et al. 1999) (Wilson et al. 1997) (Tarr & Fleischer 1999) (Hänfling & Brandl 1998) (Kontula & Vainola 2001) (Hanfling et al. 2002) (McGlashan & Hughes 2000) (Collin 2001) 5 1 mtDNA outcrossing regional regional Fst 0.872 0.07 (Collin 2001) 3 3 5 15 3 11 14 7 4 6 3 1 1 1 1 1 1 7 4 196 196 196 mtDNA mtDNA mtDNA mtDNA mtDNA mtDNA allozymes SSRs RAPDs RAPDs RAPDs outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional regional widespread widespread regional regional regional Fst Fst Fst Fst Φst, Nm Φst, Nm Fst Fst Fst Fst Fst 0.761 -0.074 0.221 0.24 0.05 0.97 0.17 0.056 0.338 0.264 0.363 0.16 1.76 1.58 10.00 0.02 1.22 4.21 0.49 0.70 0.44 (Collin 2001) (Collin 2001) (Collin 2001) (Ehinger et al. 2002) (Arendt & Smith 1998) (Arendt & Smith 1998) (Chevillon et al. 1998) (Fonseca et al. 2000) (Sebastiani et al. 2001) (Sebastiani et al. 2001) (Sebastiani et al. 2001) Cyclura carinata carinata 23 2 SSRs outcrossing endemic regional regional regional local species-wide species-wide local regional regional regional regional subspecieswide Fst, Nm 0.41 0.36 Cynictis penicillata 21 1 mtDNA endemic species-wide Φst 0.411 0.72 Daphnia ambigua 10 12 allozymes widespread local Gst 0.016 15.38 (Michels et al. 2001) Daphnia pulex Daphnia pulex 11 20 1 5 mtDNA allozymes outcrossing selfing and/or clonal selfing and/or clonal selfing and/or (Welch et al. in press) (Van Vuuren & Robinson 1997) regional regional regional regional Gst Gst 0.568 0.253 0.38 0.74 (Raay & Crease 1995) (Raay & Crease 1995) 13 Daphnia pulex 37 5 SSRs Daphnia pulex 20 13 allozymes Daphnia pulex Dasyurus maculatus Dasyurus maculatus 20 9 9 6 1 6 SSRs mtDNA SSRs clonal selfing and/or clonal selfing and/or clonal selfing and/or clonal outcrossing outcrossing Delphinapterus leucus 25 1 mtDNA Delphinapterus leucus 5 1 Delphinapterus leucus Dendroica petechia Dendroica petechia Diacamma cyaneiventre Diacamma cyaneiventre Diacamma cyaneiventre Diacamma cyaneiventre 24 8 7 3 3 7 7 Dicentrarchus labrax Dicentrarchus labrax Discus macclintocki regional regional Fst 0.4295 0.33 (Palsson 2000) regional regional Gst 0.325 0.52 (Pfrender et al. 2000) regional endemic endemic regional species-wide species-wide Gst Φst Φst 0.365 0.644 0.171 0.43 0.28 1.21 outcrossing widespread species-wide Φst 0.385 0.80 mtDNA outcrossing widespread regional Fst 0.33 1.02 5 6 1 8 1 8 1 SSRs SSRs mtDNA SSRs mtDNA SSRs mtDNA outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing widespread widespread widespread endemic endemic endemic endemic regional species-wide species-wide local local regional regional Fst Fst Φst Fst Fst Fst Fst 0.047 0.014 0.53 0.0195 0.33 0.26 0.75 5.07 17.61 0.44 12.57 1.02 0.71 0.17 4 14 10 6 6 1 SSRs SSRs mtDNA outcrossing outcrossing unknown regional regional narrow biregional regional regional Fst, Nm Fst Φst 0.007 0.0169 0.893 37.00 14.54 0.06 Drosophila buzzatii 5 10 outcrossing widespread species-wide Fst 0.136 1.59 Drosophila madeirensis 5 3 SSRs transposable elements (Pfrender et al. 2000) (Firestone et al. 1999) (Firestone et al. 1999) (Brown Gladden et al. 1997) (O'Corry-Crowe et al. 1997) (Brown Gladden et al. 1999) (Gibbs et al. 2000) (Milot et al. 2000) (Doums et al. 2002) (Doums et al. 2002) (Doums et al. 2002) (Doums et al. 2002) (García de León et al. 1997) (Lemaire et al. 2000) (Ross 1999) (Frydenberg et al. 2002) outcrossing endemic species-wide Φst 0.01069 23.14 Drosophila melanogaster Drosophila melanogaster 10 2 48 19 SSRs SSRs outcrossing outcrossing widespread widespread regional biregional Fst Fst 0.039 0.009 6.16 27.53 Drosophila mettleri Drosophila nigrospiracula 3 8 allozymes outcrossing endemic regional Fst 0.012 20.58 3 8 allozymes outcrossing endemic regional Fst 0.013 18.98 Drosophila pachea Drosophila simulans Drosophila teissieri 3 2 5 8 19 8 allozymes SSRs SSRs outcrossing outcrossing outcrossing endemic widespread regional regional biregional species-wide Fst Fst Fst 0.002 0.003 0.1218 124.75 83.08 1.80 Echinometra lucunter 10 1 mtDNA outcrossing widespread species-wide Fst 0.261 1.42 Echinometra vanbrunti Echinometra viridis 3 4 1 1 mtDNA mtDNA outcrossing outcrossing regional regional regional species-wide Fst Fst 0.057 0.361 8.27 0.89 (Lepetit et al. 2002) (Agis & Schlotterer 2001) (Colson 2002) (Pfeiler & Markow 2001) (Pfeiler & Markow 2001) (Pfeiler & Markow 2001) (Colson 2002) (Cobb et al. 2000) (McCartney et al. 2000) (McCartney et al. 2000) (McCartney et al. 14 Emberiza citrinella Emberiza schoeniclus Emydoidea blandingii Emys orbicularis 8 8 5 65 7 4 156 1 SSRs SSRs RAPDs mtDNA outcrossing outcrossing outcrossing outcrossing widespread widespread regional regional regional regional species-wide species-wide Fst Fst, Nm Φst Φst 0.01 0.0444 0.4505 0.89 24.75 5.40 0.30 0.06 Epinephelus marginatus 3 9 allozymes outcrossing regional regional Fst 0.214 0.92 Epinephelus marginatus Eretmodus cyanostictus Eretmodus cyanostictus 7 4 4 7 1 6 SSRs mtDNA SSRs outcrossing outcrossing outcrossing regional endemic endemic regional regional regional Fst Fst Fst 0.018 0.089 0.127 13.64 5.12 1.72 Erinaceus europaeus 8 6 SSRs outcrossing regional local Rst, Nm 0.079 2.34 Eubalaena glacialis Eucyclogobius newberryi 2 9 1 1 mtDNA mtDNA outcrossing outcrossing regional regional biregional regional Fst Φst 0.219 0.9023 1.78 0.05 Euhadra quaesita Ficedula hypoleuca Ficedula hypoleuca 27 3 3 1 7 1 mtDNA SSRs mtDNA unknown outcrossing outcrossing endemic regional regional regional regional regional Φst Fst Fst 0.84 0.017 0.4233 0.10 14.46 0.68 Formica cinerea Gadus morhua 23 5 5 1 SSRs mtDNA mixed outcrossing regional regional regional regional Fst Fst 0.111 0.0016 2.00 312.00 Gadus morhua Gadus morhua Galaxias maculatus Gasterosteus aculeatus Geochelone elephantopus Globicephala melas Gnamptogenys striatula Greya politella 9 14 7 3 17 4 33 27 1 5 1 7 10 8 8 1 mtDNA SSRs mtDNA SSRs SSRs SSRs SSRs mtDNA outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing mixed outcrossing regional regional widespread regional endemic regional widespread regional regional regional species-wide regional species-wide regional local species-wide Nst, Nm Fst, Nm Fst Fst Fst Fst Fst, Nm Nst -0.001 0.0084 0.458 0.159 0.188 0.006 0.172 0.979 1.16 29.40 0.59 1.32 1.08 41.42 .07 - .26 0.01 Gulo gulo Gulo gulo Halichoerus grypus Haptosquilla pulchella 12 3 2 24 12 15 8 1 SSRs SSRs SSRs mtDNA regional regional regional regional regional regional local regional? Fst Fst Fst, Nm Φst 0.0668 0.045 0.006 0.87 3.49 5.31 41.00 0.07 Heterocephalus glaber 15 1 mtDNA outcrossing outcrossing outcrossing outcrossing selfing and/or clonal regional species-wide Φct 0.68 0.24 Heterohyrax brucei Hirundichthys affinis Hyla arenicolor Hyla arenicolor Hyla regilla 16 6 20 12 7 8 58 1 1 40 SSRs RAPDs mtDNA mtDNA multilocus outcrossing outcrossing outcrossing outcrossing outcrossing regional widespread regional regional regional regional regional regional local local Fst Φst Φst Φst Φst 0.176 0.42 0.861 0.699 0.056 1.17 0.35 0.08 0.22 4.21 2000) (Lee et al. 2001) (Grapputo et al. 1998) (Mockford et al. 1999) (Lenk et al. 1999) (De Innocentiis et al. 2001) (De Innocentiis et al. 2001) (Ruber et al. 2001) (Ruber et al. 2001) (Becher & Griffiths 1998) (Rosenbaum et al. 2000) (Dawson et al. 2002) (Watanabe & Chiba 2001) (Haavie et al. 2000) (Haavie et al. 2000) (Goropashnaya et al. 2001) (Carr et al. 1995) (Árnason & Pálsson 1996) (Ruzzante et al. 1998) (Waters et al. 2000) (Reusch et al. 2001) (Ciofi et al. 2002) (Fullard et al. 2000) (Giraud et al. 2000) (Brown et al. 1997) (Kyle & Strobeck 2001) (Walker et al. 2001) (Allen et al. 1995) (Barber et al. 2002) (Faulkes et al. 1997) (Gerlach & Hoeck 2001) (Gomes et al. 1998) (Barber 1999) (Barber 1999) (Call et al. 1998) 15 Isodon obesulus 11 39 microsatellites RAPDs outcrossing regional regional Fst 0.208 0.95 Kobus kob Lagopus lagopus scoticus Lagopus mutus Lanius ludovicianus Larinus cynarae Lasiorhinus krefftii Lasiorhinus krefftii Lates niloticus Lepus americanus Lepus corsicanus Limosa haemastica 7 14 16 5 15 2 2 5 14 3 2 1 7 1 1 7 16 16 50 7 1 7 mtDNA SSRs mtDNA mtDNA allozymes SSRs SSRs allozymes SSRs mtDNA RAPDs outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional endemic narrow regional regional regional regional species-wide local species-wide local regional species-wide local local regional species-wide regional Kst Rst Φst Fst Fst Fst Fst Fst Fst Fst Fst, Nm 0.49 0.153 0.93 0.75 0.154 0.05 0.058 0.015 0.015 0.8 0.69 0.52 1.38 0.04 0.17 1.37 4.75 4.06 16.42 16.42 0.13 0.12 Linckia laevigata 4 1 mtDNA outcrossing widespread regional Φst, Nm 0.147 2.90 Linepithema humile Litoria fallax 10 22 8 1 SSRs mtDNA mixed outcrossing widespread regional local species-wide Fst Φst 0.003 0.985 83.08 0.01 Litoria pearsoniana Littorina saxatilis X tenebrosa Loligo forbesi 13 1 mtDNA outcrossing endemic species-wide Φst 0.69 0.22 15 7 1 7 mtDNA SSRs outcrossing outcrossing widespread regional regional regional Fst Fst 0.411 0.093 0.72 2.44 Loligo opalescens Lontra canadensis 11 7 6 9 SSRs SSRs outcrossing outcrossing regional regional regional? regional Fst Fst, Nm 0.0028 0.038 89.04 6.68 Loxodonta africana 3 1 mtDNA outcrossing regional regional Fst 0.553 0.40 Loxodonta africana 3 4 SSRs outcrossing regional regional Fst 0.013 18.98 Loxodonta africana Lycaon pictus Lycaon pictus Lycosid sp. Lymnaea truncatula 2 6 6 6 12 9 1 60 1 6 SSRs mtDNA SSRs mtDNA SSRs outcrossing outcrossing outcrossing outcrossing mixed regional regional regional regional regional biregional regional regional regional regional Gst Φst Fst Fst Fst 0.141 0.525 0.097 0.086 0.753 1.52 0.45 2.33 5.31 0.08 Macaca sylvanus Macquaria novemaculeata Macropus rufus Macrosiphoniella tanacetaria Macrotis lagotis 9 5 SSRs outcrossing regional regional Fst 0.118 1.87 10 13 1 1 mtDNA mtDNA regional regional species-wide biregional Φst Φst 0.05 0.18 9.50 2.28 9 10 9 1 SSRs mtDNA outcrossing outcrossing selfing and/or clonal outcrossing regional endemic regional species-wide Fst Φst 0.12 0.317 1.83 1.08 (Cooper 2000) (Birungi & Arctander 2000) (Piertney et al. 1998) (Holder et al. 2000) (Mundy et al. 1997) (Briese et al. 1996) (Taylor et al. 1994) (Taylor et al. 1994) (Hauser et al. 1998) (Burton et al. 2002) (Pierpaoli et al. 1999) (Haig et al. 1997) (Williams & Benzie 1997) (Krieger & Keller 2000) (James & Moritz 2000) (McGuigan et al. 1998) (Small & Gosling 2000) (Shaw et al. 1999) (Reichow & Smith 2001) (Blundell et al. 2002) (Nyakaana & Arctander 1999) (Nyakaana & Arctander 1999) (Whitehouse & Harley 2001) (Girman et al. 2001) (Girman et al. 2001) (Colgan et al. 2002) (Meunier et al. 2001) (von Segesser et al. 1999) (Jerry & Baverstock 1998) (Clegg et al. 1998) (Massonnet et al. 2002) (Moritz et al. 1997) 16 Macrotis lagotis 6 9 SSRs outcrossing endemic species-wide Fst 0.11 2.02 Makaira nigricans 2 1 mtDNA outcrossing widespread biregional Nm NA 1.88 Makaira nigricans 2 5 SSRs outcrossing widespread biregional Nm NA 0.74 Makaira nigricans 2 4 allozymes outcrossing widespread biregional Nm NA 1.50 Makaira nigricans Manorina melanophrys Martes pennanti Matsucoccus feytaudi Megaptera novaengliae Megaptera novaengliae 2 8 13 22 14 8 4 5 13 1 1 1 outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing widespread endemic regional regional widespread widespread biregional local species-wide species-wide species-wide regional Nm Fst Fst Nst Gst, Nm Φst NA 0.08 0.136 0.87 0.42 0.51 1.29 2.88 1.59 0.07 0.69 0.48 Megaptera novaengliae Melanochromis auratus Melanoplus oregonensis Meleagris gallopavo Meleagris gallopavo Meleagris gallopavo 8 9 15 5 5 5 5 4 1 1 83 5 scnDNA SSRs SSRs mtDNA mtDNA mtDNA SSRs + actin intron SSRs mtDNA mtDNA AFLP SSRs outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing widespread endemic regional regional regional regional regional local species-wide species-wide species-wide species-wide Fst Fst Fst Fst Fst Fst 0.048 0.151 0.46 0.546 0.175 0.113 4.96 1.41 0.59 0.42 1.18 1.96 Melitaea didyma Melospiza melodia Merluccius merluccius Merluccius merluccius Mesomys hispidus Microstomus pacificus 9 18 6 2 4 6 25 1 6 5 1 1 allozymes mtDNA SSRs SSRs mtDNA mtDNA outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional widespread regional regional regional regional regional species-wide species-wide biregional regional species-wide Fst, Nm Φst Fst Fst Fst, Nm Φst 0.09 0.294 0.039 0.0067 0.737 0.278 2.15 1.20 6.16 37.06 0.56 1.30 Microtus oeconomus 9 8 SSRs outcrossing regional regional Fst 0.167 1.25 Mogurnda adspersa Mus musculus domesticus Mus musculus musculus Myotis bechsteinii Myotis bechsteinii Myotis bechsteinii Myotis bechsteinii Myotis myotis Myotis myotis Myrmecobius fasciatus Mytilocypris ambiguosa Mytilocypris henricae Mytilocypris mytiloides 13 6 10 10 10 10 10 2 2 2 19 7 41 1 6 5 1 7 1 1 6 1 1 6 6 6 mtDNA SSRs SSRs mtDNA SSRs mtSSR mtSSR SSRs mtDNA mtDNA allozymes allozymes allozymes outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional regional regional widespread widespread narrow regional endemic regional local local local local local local local biregional biregional species-wide regional regional regional Φst Fst Fst Fst Fst Fst Fst Φst Φst Fst Fst Fst Fst 0.977 0.18 0.14 0.68 0.015 0.658 0.961 0.333 0.991 0.15 0.354 0.108 0.363 0.01 1.14 1.54 0.24 16.42 0.13 0.01 0.50 0.00 2.83 0.46 2.06 0.44 (Moritz et al. 1997) (Buonaccorsi et al. 2001) (Buonaccorsi et al. 2001) (Buonaccorsi et al. 2001) (Buonaccorsi et al. 2001) (Painter et al. 2000) (Kyle et al. 2001) (Burban et al. 1999) (Baker et al. 1994) (Baker et al. 1998) (Baker et al. 1998) (Markert et al. 1999) (Knowles 2001) (Mock et al. 2002) (Mock et al. 2002) (Mock et al. 2002) (Johannesen et al. 1996) (Fry & Zink 1998) (Lundy et al. 1999) (Lundy et al. 2000) (Patton et al. 1996) (Stepien 1999) (Van de Zande et al. 2000) (Hurwood & Hughes 1998) (Dallas et al. 1995) (Dallas et al. 1995) (Kerth et al. 2000) (Kerth et al. 2002) (Kerth et al. 2002) (Kerth et al. 2002) (Castella et al. 2000) (Castella et al. 2000) (Fumagalli et al. 1999) (Finston 2002) (Finston 2002) (Finston 2002) 17 Mytilocypris praenuncia Mytilocypris splendida 10 7 6 6 allozymes allozymes outcrossing outcrossing endemic endemic regional regional Fst Fst 0.11 0.192 2.02 1.05 Mytilus galloproviancialis 3 1 F mtDNA outcrossing regional regional Fst 0.176 1.17 Mytilus galloproviancialis Necator americanus Negaprion brevirostris 3 4 4 1 1 4 M mtDNA mtDNA SSRs regional widespread regional regional regional regional Fst Fst Fst, Nm 0.402 0.24 0.016 0.37 1.58 15.40 Nematostella vectensis Nerodia sipedon Noturus exilis Odobenus rosmarus rosmarus Odobenus rosmarus rosmarus 9 3 21 31 8 1 RAPDs SSRs mtDNA outcrossing outcrossing outcrossing selfing and/or clonal outcrossing outcrossing (Finston 2002) (Finston 2002) (Ladoukakis et al. 2002) (Ladoukakis et al. 2002) (Hawdon et al. 2001) (Feldheim et al. 2001) regional regional endemic regional local species-wide Φst Fst Fst 0.2176 0.006 0.512 0.90 41.42 0.48 (Pearson et al. 2002) (Prosser et al. 1999) (Hardy et al. 2002) 4 11 SSRs outcrossing widespread regional Fst 0.037 6.51 (Anderson et al. 1998) 4 1 mtDNA outcrossing widespread regional Φst 0.873 0.07 Odontesthes argentinensis 6 9 SSRs outcrossing regional species-wide Fst 0.029 8.37 Odontesthes argentinensis Oedura reticulata Oedura reticulata Oedura reticulata Oligoryzomys microtis Oncorhynchus clarki clarki 6 4 2 3 3 1 1 1 1 1 mtDNA mtDNA mtDNA mtDNA mtDNA outcrossing outcrossing outcrossing outcrossing outcrossing regional narrow narrow narrow regional species-wide local local local regional Φst Fst, Nm Fst, Nm Fst, Nm Fst, Nm 0.085 0.12 0.11 0.32 0.206 5.38 3.67 4.05 1.06 2.57 (Anderson et al. 1998) (Beheregaray & Sunnucks 2001) (Beheregaray & Sunnucks 2001) (Sarre 1995) (Sarre 1995) (Sarre 1995) (Patton et al. 1996) 13 6 SSRs outcrossing regional local Fst 0.121 1.82 Oncorhynchus gorbuscha 9 1 mtDNA outcrossing regional regional Φst 0.453 0.60 Oncorhynchus gorbuscha Oncorhynchus kisutch Oncorhynchus mykiss Oncorhynchus mykiss Oncorhynchus mykiss stonei Oncorhynchus nerka Oncorhynchus nerka Oncorhynchus tshawytscha Oncorhynchus tshawytscha 9 16 30 30 1 3 6 1 mtDNA SSRs scnDNA mtDNA outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional local species-wide species-wide Φst Fst Φst Φst 0.106 0.058 0.44 0.74 4.22 4.06 0.32 0.18 (Wenburg et al. 1998) (Churikov & Gharrett 2002) (Churikov & Gharrett 2002) (Small et al. 1998) (Bagley & Gall 1998) (Bagley & Gall 1998) 15 5 5 11 1 2 outcrossing outcrossing outcrossing endemic regional regional regional local local Fst Φct Fst 0.02 0.023 0.01 12.25 21.24 24.75 (Nielsen et al. 1999) (Taylor et al. 1997) (Taylor et al. 1997) 4 1 SSRs mtDNA minisatellites somatolactin intron outcrossing regional regional Fst 0.24 0.79 (Ford 2000) 4 1 outcrossing regional regional Fst 0.057 4.14 (Ford 2000) 4 12 1 1 p53 intron transferrin (noncharge changing sites) mtDNA outcrossing outcrossing regional widespread regional regional Fst Φst, Nm 0.07 0.08 3.32 6.00 (Ford 2000) (Fuller et al. 1997) Oncorhynchus tshawytscha Oryctoloagus cuniculus 18 Oryzomys capito Osmerus mordax Otis tarda Ovis canadensis Ozotoceros bezoarticus Panthera onca Panthera onca Panthera onca Panthera onca Panthera onca Panthera onca 3 49 2 23 6 4 4 3 3 2 2 1 1 1 1 1 29 1 29 1 29 1 mtDNA mtDNA mtDNA mtDNA mtDNA SSRs mtDNA SSRs mtDNA SSRs mtDNA outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional regional regional regional regional regional regional regional species-wide biregional regional species-wide species-wide species-wide triregional triregional biregional biregional Fst, Nm Gst Φst Fst Fst, Nm Fst Fst Fst Fst Fst Fst 0.012 0.96 0.49 0.497 0.37 0.063 0.3 0.295 0.065 0.342 0.045 17.34 0.02 0.52 0.51 0.87 3.72 1.17 0.60 7.19 0.48 10.61 Panthera pardus 9 1 mtDNA outcrossing widespread regional Fst 0.782 0.14 Panthera pardus Partula suturalis Partula taeniata Peltoperla tarteri Penaeus monodon Perameles gunnii Perca fluviatilis Perca fluviatilis 9 11 14 8 13 2 66 35 25 1 1 1 1 2 1 19 SSRs mtDNA mtDNA mtDNA mtDNA minisatellites mtDNA RAPDs outcrossing unknown unknown outcrossing outcrossing outcrossing outcrossing outcrossing widespread endemic endemic regional widespread endemic widespread widespread species-wide regional regional local regional local regional regional Fst Φst Φst Φst Φst Fst Φst Φst 0.364 0.682 0.474 0.17 0.835 0.352 0.66 0.48 0.44 0.23 0.55 2.44 0.10 0.46 0.26 0.27 Perna canaliculus 22 1 mtDNA outcrossing endemic species-wide Φst 0.162 2.59 Peromyscus leucopus Peromyscus maniculatus Peromyscus maniculatus Peromyscus maniculatus Petrogale lateralis Petrogale xanthopus Petrogale xanthopus Phalacrocorax carbo Phalaropus lobatus Phascolarctos cinereus Phascolarctos cinereus Phascolarctos cinereus Phocoena phocoena Phocoena phocoena 11 2 6 9 5 3 3 21 3 10 16 5 4 4 8 12 12 12 11 4 1 7 9 6 1 1 1 8 SSRs RAPDs RAPDs RAPDs SSRs SSRs mtDNA SSRs RAPDs SSRs mtDNA mtDNA mtDNA SSRs outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional regional regional widespread widespread regional regional regional widespread widespread local regional biregional local local local local regional regional species-wide species-wide regional regional regional Fst Fst, Nm Fst, Nm Fst, Nm Fst Nst Nst Rst Fst, Nm Fst Φst Fst Fst Fst 0.04 0.01 0.36 0.1 0.192 0.107 0.272 0.069 0.1 0.085 0.756 0.233 0.034 0.18 6.00 25.30 0.40 2.25 1.05 2.09 1.34 3.37 2.37 2.69 0.16 1.65 14.21 1.14 Phocoenoids dalli 4 1 mtDNA outcrossing widespread biregional Gst 0.1632 2.56 Phocoenoids dalli Phocoenoids dalli 11 11 1 6 mtDNA SSRs outcrossing outcrossing widespread widespread species-wide species-wide Φst Fst 0.0369 0.007 13.05 35.46 (Patton et al. 1996) (Bernatchez 1997) (Pitra et al. 2000) (Ramey II 1995) (González et al. 1998) (Eizirik et al. 2001) (Eizirik et al. 2001) (Eizirik et al. 2001) (Eizirik et al. 2001) (Eizirik et al. 2001) (Eizirik et al. 2001) (Uphyrkina et al. 2001) (Uphyrkina et al. 2001) (Goodacre 2002) (Goodacre 2002) (Schultheis et al. 2002) (Benzie et al. 2002) (Robinson et al. 1993) (Nesbo et al. 1999) (Nesbo et al. 1999) (Apte & Gardner 2002) (Mossman & Waser 1999) (Vucetich et al. 2001) (Vucetich et al. 2001) (Vucetich et al. 2001) (Eldridge et al. 2001) (Pope et al. 1996) (Pope et al. 1996) (Goostrey et al. 1998) (Haig et al. 1997) (Houlden et al. 1996) (Houlden et al. 1999) (Fowler et al. 2000) (Rosel et al. 1999) (Rosel et al. 1999) (McMillan & Bermingham 1996) (Escorza-Trevino & Dizon 2000) (Escorza-Trevino & 19 Dizon 2000) (Toline & Baker 1995) (Kim et al. 1998) (Haig et al. 1994) (Stangel et al. 1992) (Haig et al. 1996) (Rodriguez-Lanetty & Hoegh-Guldberg 2002) (Hoarau et al. 2002) (Castilla et al. 1998) (Gerlach & Hoeck 2001) (Emerson & Wallis 1994) (Emerson & Wallis 1994) (Rico & Turner 2002) (Roeder et al. 2001) (Dallimer et al. 2002) (Nusser et al. 1996) Phoxinus eos Phyllostis xanthopygus Picoides borealis Picoides borealis Picoides borealis 18 10 14 26 20 1 1 47 16 47 mtDNA mtDNA RAPDs allozymes RAPDs outcrossing outcrossing outcrossing outcrossing outcrossing widespread endemic endemic endemic endemic local regional species-wide species-wide species-wide Gst, Nm Φst Fst, Nm Fst, Nm Fst 0.431 0.28 0.19 0.14 0.21 0.57 1.29 1.04 1.53 0.94 Plesiastrea versipora Pleuronectes platessa Podarcis atrata 7 11 4 1 6 1 rDNA SSRs mtDNA mixed outcrossing outcrossing regional regional endemic regional regional species-wide Fst Fst Fst 0.536 0.0086 0.637 0.22 28.82 0.28 Procavia johnstoni 12 3 SSRs outcrossing regional regional Fst 0.388 0.39 Prodontria bicolorata 7 7 allozymes outcrossing endemic species-wide Fst 0.325 0.52 Prodontria modesta Pseudotropheus callainos Pygoscelis adeliae Quela quela Rallus longirostris levipes 7 3 13 8 4 7 6 5 6 16 outcrossing outcrossing outcrossing outcrossing outcrossing endemic endemic regional widespread endemic species-wide local species-wide regional species-wide Fst Fst Fst, Nm Fst Gst 0.204 0.0343 0.0007 0.002 0.28 0.98 7.04 9.00 124.75 0.64 Rana luteiventris 7 44 allozymes SSRs SSRs SSRs RAPDs multilocus microsatellites outcrossing regional local Φst 0.063 3.72 Rana sylvatica Rangifer taarandus platyrhynchus Rangifer taarandus platyrhynchus 11 5 SSRs outcrossing regional local Fst 0.014 17.61 (Call et al. 1998) (Newman & Squire 2001) 2 9 SSRs outcrossing regional local Fst 0.034 7.10 (Cote et al. 2002) 2 9 SSRs outcrossing regional local Fst 0.069 3.37 Rattus fuscipes greyii Rhinolophus ferrumequinum 16 1 MHC outcrossing regional local Fst 0.78 0.07 (Cote et al. 2002) (Seddon & Baverstock 1999) 9 6 SSRs outcrossing regional regional Fst 0.11664 1.89 Rhopalosiphum padi 11 1 mtDNA widespread widespread Cpt 0.1318 3.29 Rhopalosiphum padi Rhopalosiphum padi 5 8 7 7 SSRs SSRs widespread widespread regional regional Fst Fst 0.0603 0.0323 3.90 7.49 (Delmotte et al. 2002) (Delmotte et al. 2002) Rhopalosiphum padi Rhopalosiphum padi Rhytidoponera sp. 12 Salmo marmoratus Salmo salar Salmo salar Salmo salar 5 8 6 8 4 4 16 4 4 1 15 7 1 8 allozymes allozymes mtDNA SSRs SSRs mtDNA SSRs mixed selfing and/or clonal outcrossing selfing and/or clonal outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing (Rossiter et al. 2000) (Martínez-Torres et al. 1996) widespread widespread regional narrow regional regional widespread regional regional regional species-wide local local regional Fst Fst Fst Fst Φst Φst Fst 0.169 0.0264 0.5 0.66 0.142 0.398 0.054 1.23 9.22 0.50 0.13 1.51 0.76 4.38 (Delmotte et al. 2002) (Delmotte et al. 2002) (Tek Tay et al. 1997) (Fumagalli et al. 2002) (Tessier et al. 1997) (Tessier et al. 1997) (McConnell et al. 20 Salmo salar Salmo salar 4 14 7 5 SSRs SSRs outcrossing outcrossing regional regional local local Fst Fst 0.0957 0.034 2.36 7.10 Salmo trutta 11 38 allozymes outcrossing regional regional Fst 0.478 0.27 Salmo trutta 10 1 mtDNA outcrossing regional regional Fst 0.59 0.35 Salmo trutta Salmo trutta Salmo trutta Salmo trutta Salmo trutta 13 11 11 41 7 1 6 8 33 1 mtDNA SSRs allozymes allozymes mtDNA outcrossing outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional species-wide local local regional regional Nst Fst Fst Gst Φst 0.91 0.263 0.258 0.268 .390 - .475 0.05 0.70 0.72 0.68 - Salmo trutta Salmo trutta 58 39 1 34 mtDNA allozymes outcrossing outcrossing regional regional regional regional Φst Gst 0.559 0.455 0.39 0.30 Salmo trutta Salmo trutta Salmo trutta Salvelinus alpinus Salvelinus confluentus 9 32 5 15 47 10 7 8 6 1 SSRs SSRs SSRs SSRs mtDNA outcrossing outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional regional local regional local species-wide Fst Fst Fst, Nm Φst Φst 0.401 0.049 0.037 0.37 0.8805 0.37 4.85 6.50 0.43 0.07 Salvelinus fontinalis Salvelinus fontinalis Sceloporus woodi Schistosoma mansoni Schistosoma mansoni Schistosoma mansoni Schistosoma mansoni Sciurus vulgaris Sebastes fasciatus Sebastes fasciatus Sebastes marinus Sebastes mentella Sebastes mentella Sebastes viviparus Semibalanus balanoides Sigmodon hispidus 58 24 5 17 17 17 17 15 2 6 2 2 10 2 14 13 1 6 1 7 7 7 7 1 8 8 8 8 8 8 4 1 mtDNA SSRs mtDNA SSRs SSRs RAPDs RAPDs mtDNA SSRs SSRs SSRs SSRs SSRs SSRs SSRs MHC outcrossing outcrossing outcrossing mixed mixed mixed mixed outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing outcrossing regional regional endemic widespread widespread widespread widespread widespread regional regional regional regional regional regional widespread widespread local local species-wide local local local local regional biregional regional biregional biregional regional biregional regional regional Fst Φst Φst Fst Fst Fst Fst Φst Fst Fst Fst Fst Fst Fst Φst Fst 0.47 0.191 0.896 0.07 0.035 0.143 0.072 0.681 0.022 0.009 0.009 0.009 0.008 0.016 0.011 0.163 0.56 1.06 0.06 3.32 6.89 1.50 3.22 0.23 11.11 27.53 27.53 27.53 31.00 15.38 22.48 1.28 Silurus glanis Sistrurus c. catenatus Sitobion avenae 13 5 8 10 6 5 SSRs SSRs SSRs outcrossing outcrossing selfing and/or regional regional regional species-wide local regional Fst Fst Fst 0.364 0.164 0.032 0.44 1.27 7.56 1997) (Tessier & Bernatchez 1999) (Garant et al. 2000) (Bernatchez & Osinov 1995) (Bernatchez & Osinov 1995) (Apostolidis et al. 1997) (Estoup et al. 1998) (Estoup et al. 1998) (Bouza et al. 1999) (Weiss et al. 2000) (Machordom et al. 2000) (Bouza et al. 2001) (Aurelle & Berrebi 2001) (Ruzzante et al. 2001) (Hansen et al. 2002) (Brunner et al. 1998) (Taylor et al. 1999) (Danzmann & Ihssen 1995) (Hebert et al. 2000) (Clark et al. 1999) (Prugnolle et al. 2002) (Prugnolle et al. 2002) (Prugnolle et al. 2002) (Prugnolle et al. 2002) (Barratt et al. 1999) (Roques et al. 1999) (Roques et al. 2001) (Roques et al. 1999) (Roques et al. 1999) (Roques et al. 2001) (Roques et al. 1999) (Dufresne et al. 2002) (Pfau et al. 2001) (Triantafyllidis et al. 2002) (Gibbs et al. 1997) (Simon et al. 1999) 21 clonal Sorex araneus 17 7 Sorex araneus 14 Sorex araneus (Lugon-Moulin et al. 1999) (Lugon-Moulin & Hausser 2002) (Lugon-Moulin & Hausser 2002) (Maldonado et al. 2001) (Finch & Lambert 1996) (Garcia-Martinez et al. 1999) (Stepien & Faber 1998) (Simonsen et al. 1998) (Simonsen et al. 1998) (van Hooft et al. 2000) outcrossing regional local Fst 0.076 3.04 1 SSRs Y chromosome SSR outcrossing regional local Rst 0.738 0.09 14 8 SSRs outcrossing regional local Rst 0.061 3.85 Sorex ornatus 12 30 allozymes outcrossing endemic species-wide Fst 0.278 0.65 Sphenodon punctatus 4 135 minisatellites outcrossing endemic species-wide Fst 0.15 1.42 Stenella coeruleoalba 2 1 mtDNA outcrossing widespread biregional Gst, Nm 0.34 0.98 Stizostedion vitreum Syncerus caffer Syncerus caffer Syncerus caffer 6 11 11 9 1 1 6 14 outcrossing outcrossing outcrossing outcrossing regional regional regional regional regional regional regional species-wide Φst Kst Fst Fst 0.153 0.1 0.085 0.059 2.77 4.50 2.69 3.99 Syncerus caffer caffer 4 1 mtDNA mtDNA SSRs SSRs MHC class II DRB gene outcrossing regional regional Gst 0.076 3.04 Tamias amoenus Tanganicodus irsacae Tanganicodus irsacae Telespiza cantans 3 3 3 4 7 1 6 9 SSRs mtDNA SSRs SSRs outcrossing outcrossing outcrossing outcrossing regional endemic endemic endemic local regional regional species-wide Fst Fst Fst Fst 0.036 0.526 0.24 0.169 6.69 0.45 0.79 1.23 Tetranychus uricae 7 5 allozymes mixed widespread regional Fst 0.38 0.41 Tetrao urogallus Thalassarche chrysostoma Thalassarche chrysostoma Thalassarche melanophris Thalassarche melanophris Thaleichthys pacificus Thaumetopoea pityocampa Thaumetopoea pityocampa Thymallus arcticus Thymallus thymallus 18 10 SSRs outcrossing regional regional Fst 0.046 5.18 (Wenink et al. 1998) (Schulte-Hostedde et al. 2001) (Ruber et al. 2001) (Ruber et al. 2001) (Tarr et al. 1998) (Tsagkarakou et al. 1998) (Segelbacher & Storch 2002) 5 1 mtDNA outcrossing widespread species-wide Fst 0.00136 367.15 (Burg & Croxall 2001) 4 5 4 12 7 1 7 4 SSRs mtDNA SSRs mtDNA outcrossing outcrossing outcrossing outcrossing widespread widespread widespread regional species-wide species-wide species-wide regional Fst Fst Fst Fst 0.02 0.275 0.032 0.023 12.25 1.32 7.56 21.24 (Burg & Croxall 2001) (Burg & Croxall 2001) (Burg & Croxall 2001) (McLean et al. 1999) 8 183 AFLP outcrossing regional regional Fst, Nm 0.356 0.47 (Salvato et al. 2002) 8 6 28 1 10 1 mtDNA SSRs mtDNA outcrossing outcrossing outcrossing regional widespread regional regional regional species-wide Fst Fst Φct 0.446 0.23 0.59 0.62 0.84 0.35 Tragelaphus trepsiceros Trichechus manatus 8 8 1 1 mtDNA mtDNA outcrossing outcrossing regional regional species-wide species-wide Kst Φst 0.527 0.798 0.45 0.13 (Salvato et al. 2002) (Koskinen et al. 2002) (Koskinen et al. 2000) (Nersting & Arctander 2001) (Garcia-Rodriguez et 22 Triturus cristatus Triturus marmoratus Uria aalge Uria lomvia Urophora cardui 2 3 10 7 41 8 8 1 1 18 SSRs SSRs mtDNA mtDNA allozymes outcrossing outcrossing outcrossing outcrossing outcrossing regional regional widespread widespread regional local local biregional biregional species-wide Fst Fst Gst, Nm Gst, Nm Fst 0.0455 0.13 0.65 0.56 0.128 5.24 1.67 0.60 0.10 1.70 Ursus americanus Ursus arctos Ursus maritimus 12 4 16 10 19 16 SSRs SSRs SSRs outcrossing outcrossing outcrossing regional regional widespread regional species-wide species-wide Fst Fst Fst, Nm 0.106 0.069 0.0028 2.11 3.37 89.00 Varanus komodoensis 4 10 SSRs outcrossing endemic regional Fst, Nm 0.32 0.30 Vespula germanica Vulpes vulpes Wilsonia pusilla Zacco pachycephalus 4 4 9 12 3 7 1 26 SSRs SSRs mtDNA allozymes mixed outcrossing outcrossing outcrossing widespread regional regional endemic regional local species-wide species-wide Fst Fst Φst Fst 0.28 0.01-0.15 0.55 0.497 0.64 0.41 0.25 al. 1998) (Jehle et al. 2001) (Jehle et al. 2001) (Friesen et al. 1996) (Friesen et al. 1996) (Eber & Brandl 1997) (Marshall & Ritland 2002) (Waits et al. 2000) (Paetkau et al. 1999) (Ciofi & Bruford 1999) (Goodisman et al. 2001) (Lade et al. 1996) (Kimura et al. 2002) (Wang et al. 1999) 23 TABLE S4. Selection gradients and differentials for studies with experimentally enhanced phenotypic variation. Studies are classified according to experimental design, whether traits were varied by phenotypic manipulation or by combining varying strains, and fitness measure. Linear selection gradient or differential, with significance levels, and sample size are provided. Taxonomic group Experimental design Source of phenotypic variation Experimental conditions Trait or conditions varied or manipulated Trait class Trait name Episode, site, or treatment Fitness measure Linear gradient (β) P(β) Linear differential (s) P(s) N Source FUNGI Cochliobolus carbonum Cochliobolus carbonum Cochliobulus heterostrophus Cochliobulus heterostrophus Cochliobulus heterostrophus Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis common garden common garden common garden common garden common garden common garden common garden common garden common garden common garden common garden common garden varying strains varying strains manipulated trait manipulated trait manipulated trait varying strains varying strains varying strains varying strains varying strains varying strains varying strains laboratory among subspecies physiology against race O growth rate 0.58 na laboratory among subspecies physiology growth rate 0.17 na laboratory single allele life history growth rate 0.025 na laboratory single allele physiology growth rate 0.302 na laboratory single allele physiology against race 3 Mating type allele Against O allele Selection against T allele growth rate 0.12 na physiology competitive ability physiology competitive ability physiology competitive ability physiology competitive ability physiology competitive ability laboratory laboratory laboratory laboratory laboratory among races among races among races among races among races laboratory among races physiology laboratory among races physiology competitive ability competitive ability (Welz & Leonard 1993) (Welz & Leonard 1993) (Leonard 1977) (Leonard 1977) (Leonard 1977) (Ohara & Brown 1996) low density growth rate -0.374 na (Ohara & Brown 1996) low density growth rate -0.196 na (Ohara & Brown 1996) low density growth rate -4.62 na (Ohara & Brown 1996) low density growth rate -0.749 na (Ohara & Brown 1996) low density growth rate 0.048 na (Ohara & Brown 1996) low density growth rate -0.608 na high density growth rate 0.083 na (Ohara & Brown 1996) 24 tritici Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis tritici Erysiphe graminis tritici Ophiostoma ulmi Phytophthora infestans Puccinia graminis avenae Puccinia graminis avenae Puccinia graminis tritici Puccinia graminis tritici Puccinia graminis tritici Puccinia hordei Puccinia hordei Puccinia striiformis common garden common garden common garden common garden common garden common garden common garden common garden common garden common garden varying strains varying strains varying strains varying strains varying strains varying strains varying strains manipulated trait varying strains varying strains laboratory laboratory laboratory laboratory among races among races among races among races physiology competitive ability physiology competitive ability physiology competitive ability physiology competitive ability physiology competitive ability against nonagressive subsp. competitive ability Unneccessary virulence allele Unneccessary virulence alleles competitive ability for host variety competitive ability for temperature and density Unneccessary virulence alleles competitive ability laboratory among races laboratory among subspecies physiology laboratory among races physiology laboratory single allele physiology laboratory laboratory among races among races physiology physiology physiology common garden varying strains common garden common garden common garden common garden varying strains varying strains varying strains varying strains laboratory laboratory among races among races physiology laboratory among races physiology experimental plots among races physiology laboratory among races physiology competitive ability competitive ability (Ohara & Brown 1996) high density growth rate 0.113 na (Ohara & Brown 1996) high density growth rate -3.52 na (Ohara & Brown 1996) high density growth rate 0.164 na (Ohara & Brown 1996) high density growth rate 0.527 na (Ohara & Brown 1996) high density growth rate 0.185 growth rate 0.517 (0.087) 0.622 (0.131) growth rate 0.126 (0.013) growth rate 0.163 (0.041) growth rate 0.423 (0.06) growth rate na (Brasier 1987) na na (Thurston 1961) (Leonard 1969) na (Martens 1973) na (Loegering 1951) na (Katsuya & Green 1967) growth rate 0.279 (0.035) growth rate 0.384 (0.024) growth rate 0.194 (0.02) growth rate growth rate 0.804 0.435 (0.033) na (Osoro & Green 1976) na na na na (FalahatiRastegar et al. 1981) (FalahatiRastegar et al. 1981) (Brown & Sharp 1970) 25 Venturia inaequalis Venturia inaequalis PLANT Amphicarpaea bracteata common garden common garden manipulated trait varying strains common garden varying strains laboratory laboratory single allele among populations experimental plots among lineages physiology physiology physiology Unnecessary benomyl resistance benomyl sensitivity high/low light environments number of rosette leaves (i.e. developmental stage of flowering) (McGee & Zuck 1981) growth rate growth rate offspring after 3 generations reciprocal transplant varying strains experimental plots shading regimes Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes life history date of bolting 1996-1997, AS Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes morphology rosette diameter 1996-1997, AS Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes morphology number of rosette leaves 1997-1998, AS Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes life history date of bolting 1997-1998, AS Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes morphology rosette diameter 1997-1998, AS Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes morphology number of rosette leaves 1996-1997, SR Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes life history date of bolting 1996-1997, SR Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes morphology rosette diameter 1996-1997, SR Arabidopsis thaliana reciprocal transplant varying strains experimental plots shading regimes morphology number of rosette leaves 1997-1998, SR Arabidopsis thaliana Arabidopsis thaliana reciprocal transplant reciprocal transplant varying strains varying strains experimental plots experimental plots shading regimes shading regimes life history morphology date of bolting rosette diameter 0.57 na (McGee & Zuck 1981) na na 294 Arabidopsis thaliana morphology 0.056 0.215 (0.07) 1996-1997, AS 1997-1998, SR 1997-1998, SR total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for 0.3 <0.0001 0.59 -0.13 <0.001 -0.14 0.52 <0.0001 0.16 ns -0.18 <0.01 <.0001 <.05 294 0.69 <.0001 294 0.36 <.0001 256 -0.25 <.0001 256 0.3 <0.0001 0.41 <.0001 256 0.1 ns 0.45 <.0001 151 -0.33 <.0001 151 -0.17 <0.01 (Parker 1994) (Callahan & Pigliucci 2002) 0.54 <0.0001 0.63 <.0001 151 0.21 <0.0001 0.62 <.0001 555 -0.1 <0.001 -0.37 <.0001 555 0.56 <0.0001 0.72 <.0001 555 (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 26 Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes morphology number of rosette leaves Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes life history date of bolting Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes morphology rosette diameter Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes morphology number of rosette leaves Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes life history date of bolting Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes morphology rosette diameter unshaded early, unshaded late unshaded early, unshaded late unshaded early, unshaded late unshaded early, shaded late unshaded early, shaded late unshaded early, shaded late Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes morphology number of rosette leaves shaded early, unshaded late Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes life history date of bolting shaded early, unshaded late Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes morphology rosette diameter shaded early, unshaded late Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes morphology number of rosette leaves shaded early, shaded late Arabidopsis thaliana reciprocal transplant varying strains greenhouse shading regimes life history date of bolting shaded early, shaded late Arabidopsis thaliana Cakile edentula var. lacustris Cakile edentula var. lacustris Cakile edentula var. lacustris reciprocal transplant varying strains greenhouse shading regimes morphology rosette diameter shaded early, shaded late site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site total number of fruits/mean for site reciprocal transplant varying strains experimental plots wet and dry races physiology photosynthetic rate dry environment reproductive biomass reciprocal transplant varying strains experimental plots wet and dry races physiology stomatal conductance dry environment reproductive biomass reciprocal transplant varying strains experimental plots wet and dry races physiology water-use efficiency dry environment reproductive biomass 0.07 ns 0.12 ns 139 -0.37 <0.0001 -0.33 <.0001 139 0.48 <0.0001 0.51 <.0001 139 0.26 <0.05 0.05 ns 144 -0.61 <0.0001 -0.44 <.0001 144 0.45 <0.0001 0.59 <.0001 144 0.15 ns 0.08 0.4074 116 -0.41 <.0001 116 <.01 116 -0.45 <0.0001 0.17 ns 0.29 0.24 <0.01 -0.11 ns 127 -0.75 <0.0001 -0.62 <.0001 127 0.27 <0.0004 0.38 <.0001 127 0.41 <.05 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Callahan & Pigliucci 2002) (Dudley 1996) 57 (Dudley 1996) -0.33 <.05 57 (Dudley 1996) 0.51 <0.01 0.66 <.001 57 27 Cakile edentula var. lacustris Cakile edentula var. lacustris Cakile edentula var. lacustris Cakile edentula var. lacustris Cakile edentula var. lacustris Cakile edentula var. lacustris Cakile edentula var. lacustris reciprocal transplant varying strains experimental plots wet and dry races reciprocal transplant varying strains experimental plots wet and dry races reciprocal transplant varying strains experimental plots wet and dry races reciprocal transplant varying strains experimental plots wet and dry races reciprocal transplant varying strains experimental plots wet and dry races reciprocal transplant varying strains experimental plots wet and dry races reciprocal transplant varying strains experimental plots Impatiens capensis reciprocal transplant manipulated trait experimental plots Impatiens capensis reciprocal transplant manipulated trait experimental plots Impatiens capensis reciprocal transplant manipulated trait experimental plots Impatiens capensis reciprocal transplant manipulated trait experimental plots Impatiens capensis reciprocal transplant manipulated trait experimental plots Impatiens capensis reciprocal transplant manipulated trait experimental plots Impatiens capensis reciprocal transplant manipulated trait experimental plots Impatiens capensis Impatiens reciprocal transplant reciprocal manipulated trait manipulated experimental plots experimental wet and dry races experimentally elongated vs. nonelongated experimentally elongated vs. nonelongated experimentally elongated vs. nonelongated experimentally elongated vs. nonelongated experimentally elongated vs. nonelongated experimentally elongated vs. nonelongated experimentally elongated vs. nonelongated experimentally elongated vs. nonelongated experimentally morphology leaf size dry environment reproductive biomass morphology vegetative biomass dry environment reproductive biomass physiology photosynthetic rate wet environment reproductive biomass physiology stomatal conductance wet environment reproductive biomass physiology water-use efficiency wet environment reproductive biomass physiology leaf size wet environment reproductive biomass morphology vegetative biomass life history emergence time morphology leaf length morphology height life history emergence time morphology leaf length morphology height morphology leaf length morphology morphology height leaf length wet environment at transplantation, high density at transplantation, high density at transplantation, high density at transplantation, low density at transplantation, low density at transplantation, low density after transplantation, high density after transplantation, high density after reproductive biomass number of reproductive structures number of reproductive structures number of reproductive structures number of reproductive structures number of reproductive structures number of reproductive structures number of reproductive structures number of reproductive structures number of (Dudley 1996) 0.2 ns 0.54 <.001 57 (Dudley 1996) 0.89 <.001 57 (Dudley 1996) 0.18 <.05 99 (Dudley 1996) 0.1 ns 99 (Dudley 1996) 0.01 ns 0.01 ns 99 (Dudley 1996) 0 ns 0 ns 99 (Dudley 1996) 0.39 <.001 99 -0.139 <0.01 203 0.223 <0.001 203 0.082 ns 203 -0.078 ns 193 0.25 <0.01 193 -0.153 <0.05 193 0.129 <0.05 203 0.252 0.468 <0.001 <0.001 203 193 (Dudley & Schmitt 1996) (Dudley & Schmitt 1996) (Dudley & Schmitt 1996) (Dudley & Schmitt 1996) (Dudley & Schmitt 1996) (Dudley & Schmitt 1996) (Dudley & Schmitt 1996) (Dudley & Schmitt 1996) (Dudley & 28 capensis transplant trait plots experimental plots elongated vs. nonelongated experimentally elongated vs. nonelongated height transplantation, low density after transplantation, low density reproductive structures number of reproductive structures Impatiens capensis reciprocal transplant manipulated trait morphology Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant morphology plant size Bruuk total seeds produced 0.41 Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant life history growth habit Bruuk total seeds produced 0 ns Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant life history onset of flowering Bruuk total seeds produced 0.05 Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant morphology fresh weight Bruuk total seeds produced Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant morphology plant size Heteren Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant life history growth habit Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant life history Plantago lanceolata reciprocal transplant varying strains experimental plots reciprocal transplant Plantago lanceolata reciprocal transplant varying strains experimental plots Plantago lanceolata Plantago reciprocal transplant reciprocal varying strains varying experimental plots experimental -0.119 <0.05 <0.001 193 <.001 189 -0.14 ns 189 ns 0.07 ns 32 -0.3 ns 0.44 ns 189 total seeds produced 0.36 <0.001 0.46 <0.001 312 Heteren total seeds produced -0.19 <0.001 -0.28 <0.001 312 onset of flowering Heteren total seeds produced -0.01 ns -0.36 <0.001 283 morphology fresh weight Heteren total seeds produced -0.08 ns 0.19 <0.001 312 reciprocal transplant morphology plant size Junne total seeds produced 0.29 <0.001 0.75 <0.001 247 reciprocal transplant reciprocal life history life history growth habit onset of Junne Junne total seeds produced total seeds -0.26 -0.11 -0.06 -0.39 ns <0.05 247 140 ns ns 1.25 Schmitt 1996) (Dudley & Schmitt 1996) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (van 29 lanceolata transplant strains plots transplant Plantago lanceolata reciprocal transplant varying strains experimental plots Sinapis arvensis reciprocal transplant varying strains greenhouse Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis reciprocal transplant varying strains Sinapis arvensis ANIMAL Chelydra serpentina reciprocal transplant varying strains greenhouse reciprocal transplant lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments lineages for stress treatments markrecapture manipulated trait natural conditions hatchling size by incubation greenhouse greenhouse greenhouse greenhouse greenhouse greenhouse greenhouse greenhouse greenhouse greenhouse flowering produced morphology fresh weight Junne morphology leaf hair density control morphology leaf size morphology leaf hair density morphology leaf size morphology leaf hair density morphology leaf size morphology leaf hair density morphology leaf size morphology leaf hair density morphology leaf size morphology leaf hair density morphology leaf size control water potential of control high boron high boron high salt high salt low water low water low light low light low nutrient low nutrient total seeds produced seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean seed weight/treatment mean recapture 0.04 ns 0.16 ns 247 0.07 (0.07) ns 0.09 ns 144 -0.05 (0.07) -0.007 (0.09) 0.09 (0.09) -0.16 (0.12) 0.21 (0.11) -0.05 (0.07) 0.23 (0.07) -0.38 (0.18) 0.51 (0.19) -0.01 (0.03) Tienderen & van der Toorn 1991) (van Tienderen & van der Toorn 1991) (Roy et al. 1999) (Roy et al. 1999) ns -0.08 ns 144 (Roy et al. 1999) ns -0.004 ns 144 (Roy et al. 1999) ns 0.08 ns 144 (Roy et al. 1999) ns -0.18 ns 144 (Roy et al. 1999) ns 0.22 <0.05 144 (Roy et al. 1999) ns -0.15 <0.05 144 (Roy et al. 1999) <0.001 0.25 <0.001 144 (Roy et al. 1999) <0.001 -0.45 <0.01 144 (Roy et al. 1999) <0.001 0.52 <0.001 144 (Roy et al. 1999) ns 0.01 ns 144 (Roy et al. 1999) -0.03 (0.03) ns -0.156 ns -0.03 ns 144 112 (Janzen 1993) 30 conditions Chelydra serpentina markrecapture manipulated trait natural conditions Chelydra serpentina markrecapture manipulated trait natural conditions Chelydra serpentina markrecapture manipulated trait natural conditions Chelydra serpentina markrecapture manipulated trait natural conditions Chelydra serpentina markrecapture manipulated trait natural conditions Chelydra serpentina markrecapture manipulated trait natural conditions Chelydra serpentina markrecapture manipulated trait natural conditions hatchling size by incubation conditions hatchling size by incubation conditions hatchling size by incubation conditions hatchling size by incubation conditions hatchling size by incubation conditions hatchling size by incubation conditions hatchling size by incubation conditions Chelydra serpentina common garden manipulated trait experimental pond incubation temperature Chelydra serpentina common garden manipulated trait experimental pond incubation temperature Chelydra serpentina Chelydra serpentina Chelydra serpentina common garden common garden common garden manipulated trait manipulated trait manipulated trait experimental pond experimental pond experimental pond Parus caeruleus markrecapture manipulated trait natural conditions incubation temperature incubation temperature incubation temperature crossfostering, brood size manipulation crossfostering, brood size manipulation crossfostering, Parus caeruleus Parus caeruleus markrecapture markrecapture manipulated trait manipulated trait natural conditions natural conditions incubation substrate (Janzen 1993) life history morphology morphology morphology morphology morphology behavior morphology morphology morphology behavior clutch size PC1 (body size and shape) PC2 (body size and shape) PC3 (body size and shape) PC4 (body size and shape) PC5 (body size and shape) recapture recapture 112 0.327 <0.05 112 (Janzen 1993) recapture 0.048 ns 112 (Janzen 1993) recapture 0.282 <0.05 112 (Janzen 1993) recapture 0.162 <0.05 112 (Janzen 1993) recapture 0.057 ns 112 (Janzen 1993) locomotor performance PC1 (body size and shape) PC2 (body size and shape) PC3 (body size and shape) recapture behavior morphology nestling fledging condition combined years and treatments hatching date nestling fledging combined years and treatments 1992, combined morphology ns (Janzen 1993) running speed propensity to run control -0.096 -0.001 survival to first year -0.901 (0.884) survival to first year -0.012 (0.234) survival to first year survival to first year survival to first year -0.247 (0.262) 0.809 (1.822) 0.758 (0.376) survival to first year 0.027 (0.016) ns 112 (Janzen 1995) ns 0.295 ns 121 (Janzen 1995) ns 0.093 ns 121 (Janzen 1995) ns -0.408 ns 121 ns 0.196 ns 121 <0.05 0.671 ns 121 <0.05 (Janzen 1995) (Janzen 1995) (Merila et al. 1999) 3659 (Merila et al. 1999) survival to first year survival to first year -0.019 (0.016) 0.034 (0.032) ns 3659 ns 1038 (Merila et al. 1999) 31 Parus caeruleus Phyllonorycter mespilella Phyllonorycter mespilella Phyllonorycter mespilella markrecapture common garden common garden common garden manipulated trait manipulated trait manipulated trait manipulated trait natural conditions experimental plots experimental plots experimental plots brood size manipulation crossfostering, brood size manipulation crossfostering, brood size manipulation crossfostering, brood size manipulation crossfostering, brood size manipulation crossfostering, brood size manipulation crossfostering, brood size manipulation crossfostering, brood size manipulation timing of sapfeeding stage timing of sapfeeding stage timing of sapfeeding stage Salmo salar common garden manipulated trait experimental pond Salmo salar Thalassoma common garden disturbed manipulated trait varying experimental pond natural Parus caeruleus Parus caeruleus Parus caeruleus Parus caeruleus Parus caeruleus Parus caeruleus markrecapture markrecapture markrecapture markrecapture markrecapture markrecapture manipulated trait manipulated trait manipulated trait manipulated trait manipulated trait manipulated trait natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions condition treatments control hatching date 1992, combined treatments morphology nestling fledging condition 1993, combined treatments control hatching date 1993, combined treatments morphology nestling fledging condition 1994, combined treatments control hatching date 1994, combined treatments morphology nestling fledging condition 1995, combined treatments control life history hatching date sapfeeding duration sapfeeding duration sapfeeding duration egg size life history egg size egg size dominance life history morphology egg size standard life history life history (Merila et al. 1999) survival to first year 0.02 (0.032) ns 1038 (Merila et al. 1999) survival to first year -0.019 (0.029) ns 1177 (Merila et al. 1999) survival to first year 0.05 (0.029) ns 1177 (Merila et al. 1999) survival to first year 0.041 (0.041) ns 594 (Merila et al. 1999) survival to first year -0.001 (0.041) ns 594 (Merila et al. 1999) survival to first year 0.043 (0.034) ns 850 (Merila et al. 1999) 1995, combined treatments mixed treatments mixed treatments mixed treatments survival to first year total survivorship survivorship from parasitoid suvivorship from other -0.01 (0.034) 0.488 (0.126) 0.001 1401 1988-1990 offspring survival proportion offspring survivalX potential maternal fecundity quality of 0.358 (0.145) 0.35 0.009 <0.01 1401 70 ns 850 0.33 <0.001 0.34 <0.001 0.16 ns (McGregor 1998) (McGregor 1998) (McGregor 1998) (Einum & Fleming 2000) (Einum & Fleming 2000) (Warner & 32 bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum habitat, markrecapture disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies length mating site Schultz 1992) (Warner & Schultz 1992) morphology body depth 1988-1990 quality of mating site 0.09 ns 70 (Warner & Schultz 1992) morphology tail length 1988-1990 quality of mating site 0.08 ns 70 (Warner & Schultz 1992) morphology pectoral fin length 1988-1990 quality of mating site 0.26 <0.05 70 (Warner & Schultz 1992) morphology black + white area 1988-1990 quality of mating site 0.05 ns 70 (Warner & Schultz 1992) morphology relative white area 1988-1990 quality of mating site -0.04 ns 70 (Warner & Schultz 1992) morphology standard length 1989-1990 quality of mating site 0.64 <0.01 55 (Warner & Schultz 1992) morphology body depth 1989-1990 quality of mating site 0.13 ns 55 (Warner & Schultz 1992) morphology morphology tail length pectoral fin length 1989-1990 1989-1990 quality of mating site quality of mating site 0.14 ns 55 0.46 <0.05 55 (Warner & Schultz 33 Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat varying strains varying strains varying strains varying strains varying strains varying strains varying strains natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions Thalassoma bifasciatum disturbed habitat varying strains natural conditions Thalassoma bifasciatum disturbed habitat varying strains natural conditions through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through 1992) (Warner & Schultz 1992) morphology black + white area 1989-1990 quality of mating site 0.07 ns 55 (Warner & Schultz 1992) morphology relative white area 1989-1990 quality of mating site -0.27 ns 55 (Warner & Schultz 1992) morphology body weight 1989-1990 quality of mating site -0.1 ns 55 (Warner & Schultz 1992) physiology gut parasites 1989-1990 quality of mating site -0.07 ns 55 (Warner & Schultz 1992) physiology muscle parasites 1989-1990 quality of mating site -0.06 ns 55 (Warner & Schultz 1992) morphology standard length 1988-1990 mating site defense 0.32 <0.01 67 (Warner & Schultz 1992) morphology body depth 1988-1990 mating site defense 0.07 ns 67 (Warner & Schultz 1992) morphology tail length morphology pectoral fin length 1988-1990 mating site defense 1988-1990 mating site defense 0.09 -0.04 ns ns 67 67 (Warner & Schultz 1992) 34 Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting (Warner & Schultz 1992) morphology black + white area 1988-1990 mating site defense 0.02 ns 67 (Warner & Schultz 1992) morphology relative white area 1988-1990 mating site defense 0.18 ns 67 (Warner & Schultz 1992) morphology standard length 1989-1990 mating site defense 0.47 <0.05 55 (Warner & Schultz 1992) morphology body depth 1989-1990 mating site defense 0.23 ns 55 (Warner & Schultz 1992) morphology tail length 1989-1990 mating site defense -0.06 ns 55 (Warner & Schultz 1992) morphology pectoral fin length 1989-1990 mating site defense -0.09 ns 55 (Warner & Schultz 1992) morphology black + white area 1989-1990 mating site defense 0.02 ns 55 (Warner & Schultz 1992) morphology morphology relative white area body weight 1989-1990 1989-1990 mating site defense mating site defense 0.39 <0.05 55 (Warner & Schultz 1992) -0.29 ns 55 35 Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males (Warner & Schultz 1992) physiology gut parasites 1989-1990 mating site defense -0.02 ns 55 (Warner & Schultz 1992) physiology muscle parasites 1989-1990 mating site defense -0.07 ns 55 (Warner & Schultz 1992) morphology standard length 1988-1990 attractiveness to female 0.12 ns 63 (Warner & Schultz 1992) morphology body depth 1988-1990 attractiveness to female -0.03 ns 63 (Warner & Schultz 1992) morphology tail length 1988-1990 attractiveness to female -0.06 ns 63 (Warner & Schultz 1992) morphology pectoral fin length 1988-1990 attractiveness to female 0.11 ns 63 (Warner & Schultz 1992) morphology black + white area 1988-1990 attractiveness to female 0.07 ns 63 (Warner & Schultz 1992) morphology relative white area 1988-1990 attractiveness to female 0.19 <0.05 63 (Warner & Schultz 1992) morphology standard length 1989-1990 attractiveness to female 0.04 ns 51 36 Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thalassoma bifasciatum Thamnophis elegans disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat disturbed habitat common garden, markrecapture varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains varying strains natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions natural conditions laboratory, natural conditions dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males dominance hierarchies through transplanting males birth size (Warner & Schultz 1992) morphology body depth 1989-1990 attractiveness to female 0.06 ns 51 (Warner & Schultz 1992) morphology tail length 1989-1990 attractiveness to female 0.04 ns 51 (Warner & Schultz 1992) morphology pectoral fin length 1989-1990 attractiveness to female 0.12 ns 51 (Warner & Schultz 1992) morphology black + white area 1989-1990 attractiveness to female 0 ns 51 (Warner & Schultz 1992) morphology relative white area 1989-1990 attractiveness to female 0.28 ns 51 (Warner & Schultz 1992) morphology body weight 1989-1990 attractiveness to female -0.2 ns 51 (Warner & Schultz 1992) physiology gut parasites 1989-1990 attractiveness to female -0.11 ns 51 (Warner & Schultz 1992) physiology life history muscle parasites birth size 1989-1990 attractiveness to female survivorship 0.05 0.23 (0.04) ns 51 (Bronikowski 2000) 0.001 448 TABLE S5. Effects of quantitative trait loci for animal crosses. Species Acyrthosiphon pisum pisum Acyrthosiphon pisum pisum Acyrthosiphon pisum pisum Acyrthosiphon pisum pisum Acyrthosiphon pisum pisum Acyrthosiphon pisum pisum Acyrthosiphon pisum pisum Caenorhabditis elegans Caenorhabditis elegans Caenorhabditis elegans Caenorhabditis elegans Caenorhabditis elegans Caenorhabditis elegans Caenorhabditis elegans Caenorhabditis elegans Drosophila mauritiana X D. sechellia Drosophila mauritiana X D. simulans Drosophila mauritiana X D. simulans Drosophila mauritiana X D. simulans Drosophila mauritiana X D. simulans Drosophila mauritiana X D. simulans Drosophila mauritiana X D. simulans Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster cross intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra inter inter inter inter inter inter inter intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra Trait alfalfa race - fecundity on alfalfa alfalfa race - fecundity on clover alfalfa race - acceptance of alfalfa clover race - fecundity on alfalfa clover race - fecundity on clover clover race - acceptance of alfalfa clover race - acceptance of clover mean lifespan fertility early fertility fraction age of first reproduction population growth lifespan body length fertility female pheromones anal plate bristle number clasper bristle number sex comb tooth number fifth sternite bristle number anal plate area posterior lobe area lifespan - males lifespan - females sternopleural bristle number, 18°C, males sternopleural bristle number, 25°C, males sternopleural bristle number, 29°C, males sternopleural bristle number, 18°C, females sternopleural bristle number, 25°C, females sternopleural bristle number, 29°C, females abdominal bristle number, 18°C, males abdominal bristle number, 25°C, males abdominal bristle number, 29°C, males abdominal bristle number, 18°C, females abdominal bristle number, 25°C, females abdominal bristle number, 29°C, females male lifespan - low density male lifespan - high density female lifespan - low density Trait Type life history life history life history life history life history life history life history life history life history life history life history life history life history morphology life history physiology morphology morphology morphology morphology morphology morphology life history life history morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology life history life history life history No. QTLs 4 4 5 1 3 3 1 1 2 1 4 3 8 1 3 6 6 1 2 1 3 8 4 1 5 5 4 5 4 6 4 6 6 6 5 6 3 2 3 Largest effect size 0.8 -1.46 0.78 1.46 -1.29 24 25 23 33 46.2 53.6 -29.2 50.2 -15.9 1.22 -1.52 31.26 32.24 18.23 35 30.49 13.11 23.32 26.57 13.95 16.2 15.21 20.58 4.79 6.18 -4.2 Minor QTL2 Minor QTL3 Minor QTL4 Minor QTL5 Minor QTL6 Minor QTL7 Minor QTL8 -1.02 0.9 -0.87 0.045 0.85 -0.7 0.06 14 13 -3.5 0.61 0.11 0.06 0.12 -11.8 21.7 15 14.3 46.1 -6.9 -0.69 15.6 -14.8 -0.63 -6.9 -0.6 -4.9 -15.3 6.83 7.89 6.85 13.01 4.67 3.64 7.38 5.56 6.89 3.27 4.02 3.21 -2.89 3.45 -3.76 7.71 13.83 8.15 2.86 14.55 6.1 21.51 6.59 10.32 10.78 2.68 8.79 -1.48 9.96 9.66 8.37 16.82 11.06 7.73 2.47 19.65 3.28 12.12 3.81 6.31 7.87 12.35 0.1 0.15 -21.5 3.88 4.56 7.67 10.84 8.04 13.5 12.48 15.21 19.84 4.79 4.57 10.83 5.52 -10.5 -11.2 Minor QTL9 Minor QTL10 Minor QTL11 Minor QTL12 Minor QTL13 Minor QTL14 Minor QTL15 Minor QTL16 Minor QTL17 Minor QTL18 Minor QTL19 Method of measurement SD SD SD SD SD PVE PVE PVE %D %D %D %D %D %D SD SD PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE absolute absolute absolute Reference (Hawthorne & Via 2001) (Hawthorne & Via 2001) (Hawthorne & Via 2001) (Hawthorne & Via 2001) (Hawthorne & Via 2001) (Hawthorne & Via 2001) (Hawthorne & Via 2001) (Shook & Johnson 1999) (Shook & Johnson 1999) (Shook & Johnson 1999) (Shook & Johnson 1999) (Shook & Johnson 1999) (Ayyadevara et al. 2001) (Knight et al. 2001) (Knight et al. 2001) (Coyne & Charlesworth 1997) (True et al. 1997) (True et al. 1997) (True et al. 1997) (True et al. 1997) (True et al. 1997) (True et al. 1997) (Nuzhdin et al. 1997) (Nuzhdin et al. 1997) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Gurganus et al. 1998) (Leips & Mackay 2000) (Leips & Mackay 2000) (Leips & Mackay 2000) 38 Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila melanogaster Drosophila pseudoobscura X D. persimilis Drosophila pseudoobscura X D. persimilis Drosophila simulans X D. mauritiana Drosophila simulans X D. mauritiana Drosophila simulans X D. melanogaster Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila simulans X D. sechellia Drosophila yakuba X D. santomea Gasterosteus aculeatus Gasterosteus aculeatus intra intra intra intra intra intra intra intra intra intra intra intra intra inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter inter female lifespan - high density sex comb tooth number male lifespan female lifespan hindgrooming (autonomic) quivering (autonomic) grasping (autonomic) self-righting (autonomic) cuticular hydrocarbon profile reproductive success ovariole number courtship song - interpulse interval sternopleural bristle number - line 2 courtship song - intrapulse frequency courtship song - interpulse interval posterior lobe size/shape shape of posterior lobe female pheromones male pheromone octanoic acid resistance - males octanoic acid resistance - females posterior lobe area - sim BC posterior lobe area - sec BC adj PC1 (posterior lobe shape) - sim BC adj PC1 (posterior lobe shape) - sec BC sex comb tooth number - sim BC sex comb tooth number - sec BC testes length - sim BC testes length - sec BC cyst length - sim BC cyst length - sec BC tibia length - sim BC tibia length - sec BC testicular atrophy - sim BC testicular atrophy - sec BC absence of cysts - sim BC absence of cysts - sec BC quantity of sperm - sim BC quantity of sperm - sec BC larval hairs larval toxin resistance abdominal pigmentation number of short gill rakers lateral plate number life history morphology life history life history behavior behavior behavior behavior physiology life history life history behavior morphology behavior behavior morphology morphology physiology physiology physiology physiology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology life history life history life history life history life history life history morphology physiology morphology morphology morphology 1 2 23 23 2 2 2 2 1 2 2 3 1 2 3 9 19 5 5 4 5 11 11 9 9 4 4 7 7 3 3 5 5 3 3 4 4 3 3 1 3 3 2 2 -5.24 0.34 -0.6 -0.58 -0.73 0.51 100 -0.016 -2.98 24.9 31 30 37.3 15.9 14.9 -0.23 -0.3 0.2 0.55 0.44 -0.008 -2.71 19.4 9.7 6.8 34.1 13.6 4.4 24.4 14.7 4.5 21 25 -25.5 -36.9 63.6 27.9 53.2 -58.1 72.6 46.1 -37.3 34.1 44.5 -34.9 -0.285 -0.022 -0.453 -0.315 -1.165 -0.47 100 29 18 18 -3.5 -6.1 -1.6 -6 -13.5 -34.4 12 37 -35.8 23.8 -9.6 5.8 -2.79 -0.018 -0.401 -0.103 -0.943 0.255 5 9 -18.5 -19.7 -9.3 -7.1 15.9 -0.4 -5.3 35 -22.3 31.9 40.6 19.1 0.187 -0.007 -0.235 -0.306 -0.228 0.448 15.4 10 14.9 8.5 6.6 0.1 7 6 -7 -13.6 1.6 24.5 39.3 -16.5 35.4 -15.8 34.1 2.8 10 0.063 0.229 5.7 3.8 15.5 6.7 15.8 3.3 12 -6.5 -28.8 36.5 15.6 21.7 -19.2 34 -13.5 3.7 -3.6 38.4 -6.3 51.8 -15.3 44.7 -30.3 58.5 40.9 27.2 -33.3 10.4 2.5 -15.7 -22.1 43.1 5.5 11.8 -0.7 -14.89 -30.7 30 19.1 3.2 -9.5 -36.3 4.5 -6.7 -23.3 7.7 12.7 0.7 2.6 4.3 4.2 1.3 4.4 absolute absolute absolute absolute SD SD SD SD %D SD SD PVE PVE PVE PVE %D PVE %D %D %D %D %D %D %D %D %D %D %D %D %D %D SD SD SD SD SD SD %D %D PVE PVE (Leips & Mackay 2000) (Nuzhdin & Reiwitch 2000) (Vieira et al. 2000) (Vieira et al. 2000) (Ashton et al. 2001) (Ashton et al. 2001) (Ashton et al. 2001) (Ashton et al. 2001) (Takahashi et al. 2001) (Wayne et al. 2001) (Wayne et al. 2001) (Gleason et al. 2002) (Robin et al. 2002) (Williams et al. 2001) (Williams et al. 2001) (Liu et al. 1996) (Zeng et al. 2000) (Coyne 1996b) (Coyne 1996a) (Jones 1998) (Jones 1998) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Macdonald & Goldstein 1999) (Sucena & Stern 2000) (Jones 2001) (Llopart et al. 2002) (Peichel et al. 2001) (Peichel et al. 2001) 39 Gasterosteus aculeatus Gasterosteus aculeatus Gasterosteus aculeatus Laupala kohalensis X L. paranigra Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus inter inter inter inter intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra intra dorsal spine 1 dorsal spine 2 pelvic spine length singing pulse rate variation weight at 1 week weight at 2 weeks weight at 3 weeks weight at 4 weeks weight at 5 weeks weight at 6 weeks weight at 7 weeks weight at 8 weeks weight at 9 weeks weight at 10 weeks early growth middle growth six-week growth late growth body size coronoid height superior condylar length condylar width inferior condylar length condylar base length posterior angular height posterior angular length anterior angular length superior angular length posterior corpus height coronoid base length posterior-inferior basal length anterior-inferior basal length inferior incisor alveolus length incisor alveolus width superior incisor alveolus length anterior corpus height molar alveolus height superior molar alveolus length inferior molar alveolus length superior coronoid length body weight abdominal fat weight abdominal fat percentage liver weight morphology morphology morphology behavior morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology physiology physiology physiology physiology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology morphology 2 2 1 3 7 10 16 13 16 15 14 14 16 17 11 12 14 12 11 12 8 5 9 11 8 8 5 4 8 12 5 11 5 5 4 8 11 7 8 9 10 8 4 5 1.53 2.72 3.35 9 9.6 8.8 15.6 18.3 18 18 16.3 12.7 18.4 15.2 11.3 8.5 11.1 8.4 -0.28 16.5 4.1 10.5 9.3 4.9 16.4 6.6 3.6 3.9 5.9 14.1 3.8 6.1 4.4 13.3 7.6 14.5 6.5 7 7.4 9.9 23.1 18.1 10.2 24.7 1.42 2.57 6.5 5.4 4.5 6.5 1.9 2 2.7 5.2 5.1 6.3 5.3 5.3 4.8 4.1 1.9 -0.17 3.4 3.1 2.7 2.8 3.8 2 2.8 1.9 1.6 1.8 3.9 2.7 3.3 2.4 3.6 1.7 2.9 3.7 3.5 5.5 4.6 7.1 4 8.3 4.4 6.1 2.3 3.2 5.8 5.3 4 6.3 2.3 6.7 3 7.2 2.8 7.7 6.8 2.6 0.18 3.2 3.8 2.6 5.1 1.9 7 2.5 3.5 1.9 2.8 1.5 3.5 2.9 4.4 3.2 6.5 3.2 3.2 1.9 3.4 5.2 5.1 13.4 4.7 6.6 7.6 2.5 2.7 2.3 2.4 8.2 5.1 3 6.2 3.1 3.5 3.4 1.8 2.4 0.18 2.5 2.1 9.6 2.5 2 2.6 2 2.6 3.8 4.9 6.7 2.1 3.9 1.8 2.4 1.8 2.3 2.9 2.8 2.4 3.4 7 3.9 5.3 9.7 9.2 2.1 2.2 7.8 5.9 2.5 12.4 6.4 9 2.6 4.5 3.6 5.4 3.4 -0.21 9.2 3.8 2.5 4.4 2.5 3.2 3.8 2.3 3.8 8.6 10.2 8 3.1 4.7 12 10.1 16.2 12.4 2.4 3.9 6 2 -0.19 2.5 2.8 1.3 1.9 2.9 6.6 9.1 11.5 16.3 8.7 13.6 6.3 2 4.9 8.2 2.4 -0.21 5.7 1.8 2.8 4.4 13.7 8.2 12.6 3.5 10.2 1.5 10.4 1.8 2.3 10.6 2.4 0.24 2.3 1.7 1.5 12.7 1.6 12.3 17.8 3.4 3.3 4 11.7 2.9 2.4 3.4 2.4 -0.2 2 2.1 1.4 2.3 17.7 3 3.6 3.4 3.2 15 1.3 2.1 2.6 4.4 0.17 2.8 2.2 4.4 1.9 2.4 2.1 2.7 2.1 1.8 1.6 1.9 3.5 2.9 0.22 1.5 6.5 4.4 3.4 2.3 3.1 4 5.2 2.1 2.2 1.8 2.2 3.1 6.7 4.8 5 2.8 3.3 2.9 1.6 2.6 2.4 1.5 2.7 2.6 3.2 2.8 1.9 10.6 2.8 6.1 2.2 4.7 4.9 2.9 3.5 3.7 2.5 6.8 2.7 2.4 2.8 7.3 3 4.1 5.6 3.1 3.1 6.6 3.2 4.3 8.3 3.6 3.2 2.5 2.5 3.3 10.1 7.7 2.7 2.3 3.3 2.9 3.5 2.5 0.2 3.3 6.8 1.6 2.6 4.2 2.9 3.4 6.4 4.4 3.7 2.1 1.8 2 3.6 2.3 2.6 4.4 3.4 4.3 2.2 4.3 2.9 1.5 1.3 1.9 2.1 1.7 2.8 4 1.8 2.6 2.5 3.4 2.2 3.2 5.1 1.9 3.4 4.3 3.3 3 1.5 3.4 6.6 3.3 2.1 PVE PVE PVE %D PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE SD PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE (Peichel et al. 2001) (Peichel et al. 2001) (Peichel et al. 2001) (Shaw & Parsons 2002) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Cheverud et al. 1996) (Keightley et al. 1996) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Cheverud et al. 1997) (Brockmann et al. 1998) (Brockmann et al. 1998) (Brockmann et al. 1998) (Brockmann et al. 1998) 40 Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Mus musculus Nasonia giraulti X N. vitripennis Oncorhynchus mykiss Oncorhynchus mykiss Oncorhynchus mykiss intra intra intra intra intra intra intra intra intra intra intra intra intra inter intra intra intra kidney weight spleen weight growth rate left-right centroid size (mandibular) assymetry of centroid size (mandibular) unsigned assymetry of centroid size testis weight seminal vesicle weight aggression maternal performance early growth early growth (maternal effect) molar centroid size wing size time to hatch - pooled embryonic length - pooled weight at swim-up - pooled morphology morphology life history morphology morphology morphology morphology morphology behavior behavior physiology physiology morphology morphology life history morphology morphology 5 4 20 11 2 1 5 1 2 2 5 4 3 2 3 2 2 13.4 23 11 8.78 2.24 2.38 20.7 24.2 6.18 3.02 9.53 6.57 44 14.7 13.1 15.3 8.2 7.3 7.3 8.5 6.2 10.7 4.9 2.9 2.11 3.76 3.38 3.58 17.5 11.9 13.5 9.2 4.61 2.49 9.07 5.46 1.88 9.13 2.14 3 5.21 1.82 4.6 9.5 10.9 5.3 2.15 6.57 3.93 4.66 3.86 3.17 PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE PVE %D PVE PVE PVE (Brockmann et al. 1998) (Brockmann et al. 1998) (Vaughn et al. 1999) (Klingenberg et al. 2001) (Klingenberg et al. 2001) (Klingenberg et al. 2001) (Le Roy et al. 2001) (Le Roy et al. 2001) (Brodkin et al. 2002) (Peripato et al. 2002) (Wolf et al. 2002) (Wolf et al. 2002) (Workman et al. 2002) (Weston et al. 1999) (Robison et al. 2001) (Robison et al. 2001) (Robison et al. 2001) 41 References Aagaard JE, Krutovskii KV, Strauss SH (1998) RAPD markers of mitochondrial origin exhibit lower population diversity and higher differentiation than RAPDs of nuclear origin in Douglas fir. Molecular Ecology 7, 801-812. Aagaard JE, Vollmer SS, Sorensen FC, Strauss SH (1995) Mitochondrial-DNA Products among Rapd Profiles Are Frequent and Strongly Differentiated between Races of Douglas-Fir. Molecular Ecology 4, 441-446. Abbott RJ, Chapman HM, Crawford RMM, Forbes DG (1995) Molecular Diversity and Derivations of Populations of Silene- Acaulis and SaxifragaOppositifolia from the High Arctic and More Southerly Latitudes. Molecular Ecology 4, 199-207. Agis M, Schlotterer C (2001) Microsatellite variation in natural Drosophila melanogaster populations from New South Wales (Australia) and Tasmania. Molecular Ecology 10, 1197-1205. Akimoto M, Shimamoto Y, Morishima H (1998) Population genetic structure of wild rice Oryza glumaepatula distributed in the Amazon flood area influenced by its life- history traits. Molecular Ecology 7, 1371-1381. Aldrich PR, Hamrick JL, Chavarriaga P, Kochert G (1998) Microsatellite analysis of demographic genetic structure in fragmented populations of the tropical tree Symphonia globulifera. Molecular Ecology 7, 933-944. Alexandrino J, Froufe E, Arntzen JW, Ferrand N (2000) Genetic subdivision, glacial refugia and postglacial recolonization in the golden-striped salamander, Chioglossa lusitanica (Amphibia : Urodela). Molecular Ecology 9, 771-781. Allen PJ, Amos W, Pomeroy PP, Twiss SO (1995) Microsatellite variation in grey seals (Halichoerus grypus) shows evidence of genetic differentiation between two British breeding colonies. Molecular Ecology 4, 653-662. Allnutt TR, Newton AC, Lara A, et al. (1999) Genetic variation in Fitzroya cupressoides (alerce), a threatened South American conifer. Molecular Ecology 8, 975-987. Anderson LW, Born EW, Gjertz I, et al. (1998) Population structure and gene flow of the Atlantic walrus (Odobenus rosmarus rosmarus) in the eastern Atlantic Artic based on mitochondrial DNA and microsatellite variation. Molecular Ecology 7, 1323-1336. Apostolidis AP, Triantaphyllidis D, Kouvatsi A, Economidis PS (1997) Mitochondrial DNA sequence variation and phylogeography among Salmo trutta L. (Greek brown trout) populations. Molecular Ecology 6, 531-542. Apte S, Gardner JPA (2002) Population genetic subdivision in the New Zealand greenshell mussel (Perna canaliculus) inferred from single-strand conformation polymorphism analysis of mitochondrial DNA. Molecular Ecology 11, 1617-1628. Arafeh RMH, Sapir Y, Shmida A, et al. (2002) Patterns of genetic and phenotypic variation in Iris haynei and I-atrofusca (Iris sect. Oncocyclus = the royal irises) along an ecogeographical gradient in Israel and the West Bank. Molecular Ecology 11, 39-53. Arendt S, Smith MJ (1998) Genetic diversity and population structure in two species of sea cucumber: differing patterns according to mode of development. Molecular Ecology 7, 1053-1064. Árnason R, Pálsson S (1996) Mitochondrial cytochrome b DNA sequence variation of Atlantic cod Gadus morhua, from Norway. Molecular Ecology 5, 715724. Ashton K, Wagoner AP, Carrillo R, Gibson G (2001) Quantitative trait loci for the monoamine-related traits heart rate and headless behavior in Drosophila melanogaster. Genetics 157, 283-294. Auge H, Neuffer B, Erlinghagen F, Grupe R, Brandl R (2001) Demographic and random amplified polymorphic DNA analyses reveal high levels of genetic diversity in a clonal violet. Molecular Ecology 10, 1811-1819. 42 Aurelle D, Berrebi P (2001) Genetic structure of brown trout (Salmo trutta L.) populations from south-western France: data from mitochondrial control region variability. Molecular Ecology 10, 1551-1561. Ayres DR, Ryan FJ (1997) The clonal and population structure of a rare endemic plant, Wyethia reticulata (Asteraceae): allozyme and RAPD analysis. Molecular Ecology 6, 761-772. Ayyadevara S, Ayyadevara R, Hou S, Thaden JJ, Reis RJS (2001) Genetic mapping of quantitative trait loci governing longevity of Caenorhabditis elegans in recombinant-inbred progeny of a Bergerac-BO x RC301 interstrain cross. Genetics 157, 655-666. Bagley MJ, Gall GAE (1998) Mitochondrial and nuclear DNA sequence variability among populations of rainbow trout (Oncorhynchus mykiss). Molecular Ecology 7, 945-962. Baker CS, Medrano-Gonzalez L, Calambokidis J, et al. (1998) Population structure of nuclear and mitochondrial DNA variation among humback whales in the North Pacific. Molecular Ecology 7, 695-707. Baker CS, Slade RW, Bannisgter SL, et al. (1994) Hierarchical structure of mitochondrial DNA gene flow among humpback whales Megaptera novaeangliae, worldwide. Molecular Ecology 3, 313-327. Barber PH (1999) Patterns of gene flow and population genetic structure in the canyon treefrog, Hyla arenicolor (Cope). Molecular Ecology 8, 563-576. Barber PH, Palumbi SR, Erdmann MV, Moosa MK (2002) Sharp genetic breaks among populations of Haptosquilla pulchella (Stomatopoda) indicate limits to larval transport: patterns, causes, and consequences. Molecular Ecology 11, 659-674. Barratt EM, Gurnell J, Malarky G, Deaville R, Bruford MW (1999) Genetic structure of fragmented populations of red squirrel (Sciurus vulgaris) in the UK. Molecular Ecology 8, S55-S63. Bartish IV, Jeppsson N, Nybom H (1999) Population genetic structure in the dioecious pioneer plant species Hippophae rhamnoides investigated by random amplified polymorphic DNA (RAPD) markers. Molecular Ecology 8, 791-802. Becher SA, Griffiths R (1998) Genetic differentiation among local populations of the European hedgehog (Erinaceus europaeus) in mosaic habitats. Molecular Ecology 7, 1599-1604. Beheregaray LB, Sunnucks P (2001) Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Molecular Ecology 10, 2849-2866. Belahbib N, Pemonge MH, Ouassou A, et al. (2001) Frequent cytoplasmic exchanges between oak species that are not closely related: Quercus suber and Q-ilex in Morocco. Molecular Ecology 10, 2003-2012. Benzie JAH, Ballment E, Forbes AT, et al. (2002) Mitochondrial DNA variation in Indo-Pacific populations of the giant tiger prawn, Penaeus monodon. Molecular Ecology 11, 2553-2569. Bernatchez L (1997) Mitochondrial DNA analysis confirms the existence of two glacial races of rainbow smelt Osmerus mordax and their reproductive isolation in the St. Lawrence River estuary (Québec, Canada). Molecular Ecology 6, 73-83. Bernatchez L, Osinov A (1995) Genetic diversity of trout (genus Salmo) from its most eastern native range based on mitochondrial DNA and nuclear gene variation. Molecular Ecology 4, 285-297. Birungi J, Arctander P (2000) Large sequence divergence of mitochondrial DNA genotypes of the control region within populations of the African antelope, kob (Kobus kob). Molecular Ecology 9, 1997-2008. Blundell GM, Ben-David M, Groves P, Bowyer RT, Geffen E (2002) Characteristics of sex-biased dispersal and gene flow in coastal river otters: implications for natural recolonization of extirpated populations. Molecular Ecology 11, 289-303. Bonnin I, Colas B, Bacles C, et al. (2002) Population structure of an endangered species living in contrasted habitats: Parnassia palustris (Saxifragaceae). Molecular Ecology 11, 979-990. 43 Bonnin I, Ronfort J, Wozniak F, Olivieri I (2001) Spatial effects and rare outcrossing events in Medicago truncatula (Fabaceae). Molecular Ecology 10, 13711383. Bouza C, Arias J, Castro J, Sanchez L, Martinez P (1999) Genetic structure of brown trout, Salmo trutta L., at the southern limit of the distribution range of the anadromous form. Molecular Ecology 8, 1991-2001. Bouza C, Castro J, Sanchez L, Martinez P (2001) Allozymic evidence of parapatric differentiation of brown trout (Salmo trutta L.) within an Atlantic river basin of the Iberian Peninsula. Molecular Ecology 10, 1455-1469. Brasier CM (1987) Recent genetic changes in the Ophiostoma ulmi population: the threat to the future of the elm. In: Populations of plant pathogens (eds. Wolfe MS, Caten CE), pp. 213-226. Blackwell Scientific Publications, Oxford. Briese DT, Espiau C, Pouchot-Lermans A (1996) Micro-evolution in the weevil genus Larinus: the formation of host biotypes and speciation. Molecular Ecology 5, 531-545. Brockmann GA, Haley CS, Renne U, Knott SA, Schwerin M (1998) Quantitative trait loci affecting body weight and fatness from a mouse line selected for extreme high growth. Genetics 150, 369-381. Brodkin ES, Goforth SA, Keene AH, Fossella JA, Silver LM (2002) Identification of quantitative trait loci that affect aggressive behavior in mice. Journal of Neuroscience 22, 1165-1170. Bronikowski AM (2000) Experimental evidence for the adaptive evolution of growth rate in the garter snake Thamnophis elegans. Evolution 54, 1760-1767. Brown Gladden JG, Ferguson MM, Clayton JW (1997) Matriarchal genetic population structure of North American beluga whales Delphinapterus leucas (Cetacea: Monodontidae). Molecular Ecology 6, 1033-1046. Brown Gladden JG, Ferguson MM, Friesen MK, Clayton JW (1999) Population structure of North American beluga whales (Delphinapterus leucas) based on nuclear DNA microsatellite variation and contrasted with the population structure revealed by mitochondrial DNA variation. Molecular Ecology 8, 347363. Brown JF, Sharp EL (1970) The relative survival ability of pathogenic types of Puccinia striiformis in mixtures. Phytopathology 60, 529-533. Brown JM, Leebens-Mack HH, Thompson JN, Pellmyr O, Harrison RG (1997) Phylogeography and host association in a pollinating seed parasite Greya politella (Lepidoptera: Prodoxidae). Molecular Ecology 6, 215-224. Brown RP, Campos-Delgado R, Pestano A (2000) Mitochondrial DNA evolution and population history of the Tenerife skink Chalcides viridanus. Molecular Ecology 9, 1061-1067. Brunner PC, Douglas MR, Bernatchez L (1998) Microsatellite and mitochondrial DNA assessment of population structure and stocking effects in Artic charr Salvelinus alpinus (Teleostei: Salmonidae) from central alpine lakes. Molecular Ecology 7, 209-223. Brzosko E, Wróblewska A, Ratkiewicz M (2002) Spatial genetic structure and clonal diversity of island populations of lady's slipper (Cypripedium calceolus) from the Biebrza National Park (northeast Poland). Molecular Ecology 11, 2499-2509. Bucci G, Anzidei M, Madaghiele A, Vendramin GG (1998) Detection of haplotypic variation and natural hybridization in halepensis-complex pine species using chloroplast simple sequence repeat (SSR) markers. Molecular Ecology 7, 1633-1643. Bucci G, Vendramin GG (2000) Delineation of genetic zones in the European Norway spruce natural range: preliminary evidence. Molecular Ecology 9, 923934. Bucheli E, Gautschi B, Shykoff JA (2001) Differences in population structure of the anther smut fungus Microbotryum violaceum on two closely related host species, Silene latifolia and S. dioicia. Molecular Ecology 10, 285-294. Buonaccorsi VP, McDowell JR, Graves JE (2001) Reconciling patterns of inter-ocean molecular variance from four classes of molecular markers in blue marlin (Makaira nigricans). Molecular Ecology 10, 1179-1196. 44 Burban C, Petit RJ, Carcreff E, Jactel H (1999) Rangewide variation of the maritime pine bast scale Matsucoccus feytaudi Duc. (Homoptera: Matsucoccidae) in relation to the genetic structure of the host. Molecular Ecology 8, 1593-1602. Burg TM, Croxall JP (2001) Global relationships amongst black-browed and grey-headed albatrosses: analysis of population structure using mitochondrial DNA and microsatellites. Molecular Ecology 10, 2647-2660. Burt A, DeChairo BM, Koenig GL, et al. (1997) Molecular markers reveal differentiation among isolates of Coccidioides immitis from California, Arizona and Texas. Molecular Ecology 6, 781-786. Burton C, Krebs CJ, Taylor EB (2002) Population genetic structure of the cyclic snowshoe hare (Lepus americanus) in southwestern Yukon, Canada. Molecular Ecology 11, 1689-1701. Buso GSC, Rangel PH, Ferreira ME (1998) Analysis of genetic variability of South American wild rice populations (Oryza glumaepatula) with isozymes and RAPD markers. Molecular Ecology 7, 107-117. Bussell JD (1999) The distribution of random amplified polymorphic DNA (RAPD) diversity amongst populations of Isotoma petraea (Lobeliaceae). Molecular Ecology 8, 775-789. Byrne M, Macdonald B, Coates D (1999) Divergence in the chloroplast genome and nuclear rDNA of the rare Western Australian plant Lambertia orbifolia Gardner (Proteaceae). Molecular Ecology 8, 1789-1796. Call DR, Hallett JG, Mech SG, Evans M (1998) Considerations for measuring genetic variation and population structure with multilocus fingerprinting. Molecular Ecology 7, 1337-1346. Callahan HS, Pigliucci M (2002) Shade-induced plasticity and its ecological significance in wild populations of Arabidopsis thaliana. Ecology 83, 1965-1980. Cardoso MA, Provan J, Powell W, Ferreira PCG, De Oliveira DE (1998) High genetic differentiation among remnant populations of the endangered Caesalpinia echinata Lam. (Leguminosae- Caesalpinioideae). Molecular Ecology 7, 601-608. Cardoso SRS, Eloy NB, Provan J, Cardoso MA, Ferreira PCG (2000) Genetic differentiation of Euterpe edulis Mart. populations estimated by AFLP analysis. Molecular Ecology 9, 1753-1760. Carino DA, Daehler CC (1999) Genetic variation in an apomictic grass, Heteropogon contortus, in the Hawaiian Islands. Molecular Ecology 8, 2127-2132. Carlier J, Lebrun MH, Zapater MF, Dubois C, Mourichon X (1996) Genetic structure of the global population of banana black leaf streak fungus, Mycosphaerella fijiensi. Molecular Ecology 6, 499-510. Carmichael LE, Nagy JA, Larter NC, Strobeck C (2001) Prey specialization may influence patterns of gene flow in wolves of the Canadian Northwest. Molecular Ecology 10, 2787-2798. Caron H, Dumas S, Marque G, et al. (2000) Spatial and temporal distribution of chloroplast DNA polymorphism in a tropical tree species. Molecular Ecology 9, 1089-1098. Carr SM, Sneller AJ, Howse KA, Wroblewstri JS (1995) Mitochondrial DNA sequence variation and genetic stock structure of Atlantic cod (Gadus morhua) from bay and offshore locations on the Newfoundland continental shelf. Molecular Ecology 4, 79-88. Castella V, Ruedi M, Excoffier L, et al. (2000) Is the Gibraltar Strait a barrier to gene flow for the bat Myotis myotis (Chiroptera : Vespertilionidae)? Molecular Ecology 9, 1761-1772. Castilla AM, Fern V, Fernández-pedrosa, et al. (1998) Conservation genetics of insular Podarcis lizards using partial cytochrome b seuences. Molecular Ecology 7, 1407-1411. Charbonnel N, Angers B, Rasatavonjizay R, et al. (2002) The influence of mating system, demography, parasites and colonization on the population structure of Biomphalaria pfeifferi in Madagascar. Molecular Ecology 11, 2213-2228. Cheverud JM, Routman EJ, Duarte FAM, et al. (1996) Quantitative trait loci for murine growth. Genetics 142, 1305-1319. 45 Cheverud JM, Routman EJ, Irschick DJ (1997) Pleiotropic effects of individual gene loci on mandibular morphology. Evolution 51, 2006-2016. Chevillon C, Rivet Y, Raymond M, et al. (1998) Migration/selection balance and ecotypic differentiation in the mosquito Culex pipiens. Molecular Ecology 7, 197-208. Chiang TY, Chiang YC, Chen YJ, et al. (2001) Phylogeography of Kandelia candel in East Asiatic mangroves based on nucleotide variation of chloroplast and mitochondrial DNAs. Molecular Ecology 10, 2697-2710. Chiang TY, Schaal BA (1999) Phylogeography of North American populations of the moss species Hylocomium sylendens based on the nucleotide sequence of internal transcribed spacer 2 of nuclear ribosomal DNA. Molecular Ecology 8, 1037-1042. Churikov D, Gharrett AJ (2002) Comparative phylogeography of the two pink salmon broodlines: an analysis based on a mitochondrial DNA genealogy. Molecular Ecology 11, 1077-1101. Ciofi C, Bruford MW (1999) Genetic structure and gene flow among Komodo dragon populations inferred by microsatellite loci analysis. Molecular Ecology 8, S17-S30. Ciofi C, Milinkovitch MC, Gibbs JP, Caccone A, Powell JR (2002) Microsatellite analysis of genetic divergence among populations of giant Galápagos tortoises. Molecular Ecology 11, 2265-2283. Clark AM, Bowen BW, Branch LC (1999) Effects of natural habitat fragmentation on an endemic scrub lizard (Sceloporus woodi): an historical perspective based on a mitochondrial DNA gene genealogy. Molecular Ecology 8, 1093-1104. Clegg SM, Hale P, Moritz C (1998) Molecular population genetics of the red kangaroo. Molecular Ecology 7, 679-686. Cobb M, Huet M, Lachaise D, Veuille M (2000) Fragmented forests, evolving flies: molecular variation in African populations of Drosophila teissieri. Molecular Ecology 9, 1591-1597. Colgan DJ, Brown S, Major RE, et al. (2002) Population genetics of wolf spiders of fragmented habitat in the wheat belt of New South Wales. Molecular Ecology 11, 2295-2305. Collevatti RG, Grattapaglia D, Hay JD (2001) Population genetic structure of the endangered tropical tree species Caryocar brasiliense, based on variability at microsatellite loci. Molecular Ecology 10, 349-356. Collin R (2001) The effects of mode of development on phylogeography and population structure of North Atlantic Crepidula (Gastropoda : Calyptraeidae). Molecular Ecology 10, 2249-2262. Colson I (2002) Selection and gene flow between microenvironments: The case of Drosophila at Lower Nahal Oren, Mount Carmel, Israel. Molecular Ecology 11, 1311-1316. Comes HP, Abbott RJ (2000) Random amplified polymorphic DNA (RAPD) and quantitative trait analyses across a major phylogeographical break in the Mediterranean ragwort Senecio gallicus Vill. (Asteraceae). Molecular Ecology 9, 61-76. Cooper ML (2000) Random amplified polymorphic DNA analysis of southern brown bandicoot (Isoodon obesulus) populations in Western Australia reveals genetic differentiation related to environmental variables. Molecular Ecology 9, 469-479. Cote SD, Dallas JF, Marshall F, et al. (2002) Microsatellite DNA evidence for genetic drift and philopatry in Svalbard reindeer. Molecular Ecology 11, 19231930. Coyne JA (1996a) Genetics of a difference in male cuticular hydrocarbons between two sibling species, Drosophila simulans and D-sechellia. Genetics 143, 1689-1698. Coyne JA (1996b) Genetics of differences in pheromonal hydrocarbons between Drosophila melanogaster and D-simulans. Genetics 143, 353-364. Coyne JA, Charlesworth B (1997) Genetics of a pheromonal difference affecting sexual isolation between Drosophila mauritiana and D-sechellia. Genetics 145, 1015-1030. 46 Dallas JF, Dod B, Bourset P, Prager EM, Bonhomme F (1995) Population subdivision and gene flow in Danish house mice. Molecular Ecology 4, 311-320. Dallimer M, Blackburn C, Jones PJ, Pemberton JM (2002) Genetic evidence for male biased dispersal in the red-billed quelea Quelea quelea. Molecular Ecology 11, 529-533. Danzmann RG, Ihssen PE (1995) A phylogeographic survey of brook charr (Savelinus fontinalis) in Algonquin Park, Ontario based upon mitochondrial DNA variation. Molecular Ecology 4, 681-697. Dawson IK, Chalmers KJ, Waugh R, Powell W (1993) Detection and Analysis of Genetic-Variation in Hordeum- Spontaneum Populations from Israel Using Rapd Markers. Molecular Ecology 2, 151-159. Dawson IK, Powell W (1999) Genetic variation in the Afromontane tree Prunus africana, an endangered medicinal species. Molecular Ecology 8, 151-156. Dawson MN, Louie KD, Barlow M, Jacobs DK, Swift CC (2002) Comparative phylogeography of sympatric sister species, Clevelandia ios and Eucyclogobius newberryi (Teleostei, Gobiidae), across the California Transition Zone. Molecular Ecology 11, 1065-1075. Dayanandan S, Dole J, Bawa K, Kesseli R (1999) Population structure delineated with microsatellite markers in fragmented populations of a tropical tree, Carapa guianensis (Meliaceae). Molecular Ecology 8, 1585-1592. De Innocentiis S, Sola L, Cataudella S, Bentzen P (2001) Allozyme and microsatellite loci provide discordant estimates of population differentiation in the endangered dusky grouper (Epinephelus marginatus) within the Mediterranean Sea. Molecular Ecology 10, 2163-2175. De la Rua P, Galian J, Serrano J, Moritz RFA (2001) Genetic structure and distinctness of Apis mellifera L. populations from the Canary Islands. Molecular Ecology 10, 1733-1742. Delmotte F, Leterme N, Gauthier JP, Rispe C, Simon JC (2002) Genetic architecture of sexual and asexual populations of the aphid Rhopalosiphum padi based on allozyme and microsatellite markers. Molecular Ecology 11, 711-723. Deshpande AU, Apte GS, Bahulikar RA, et al. (2001) Genetic diversity across natural populations of three montane plant species from the Western Ghats, India revealed by intersimple sequence repeats. Molecular Ecology 10, 2397-2408. Despres L, Loriot S, Gaudeul M (2002) Geographic pattern of genetic variation in the European globeflower Trollius europaeus L. (Ranunculaceae) inferred from amplified fragment length polymorphism markers. Molecular Ecology 11, 2337-2347. Diaz V, Muniz LM, Ferrer E (2001) Random amplified polymorphic DNA and amplified fragment length polymorphism assessment of genetic variation in Nicaraguan populations of Pinus oocarpa. Molecular Ecology 10, 2593-2603. Dobler S, Farrell BD (1999) Host use evolution in Chrysochus milkweed beetles: evidence from behaviour, population genetics and phylogeny. Molecular Ecology 8, 1297-1307. Dodd RS, Afzal-Rafii Z, Kashani N, Budrick J (2002) Land barriers and open oceans: effects on gene diversity and population structure in Avicennia germinans L. (Avicenniaceae). Molecular Ecology 11, 1327-1338. Douhan GW, Peever TL, Murray TD (2002) Multilocus population structure of Tapesia yallundae in Washington State. Molecular Ecology 11, 2229-2239. Doums C, Cabrera H, Peeters C (2002) Population genetic structure and male-biased dispersal in the queenless ant Diacamma cyaneiventre. Molecular Ecology 11, 2251-2264. Drummond RSM, Keeling DJ, Richardson TE, Gardner RC, Wright SD (2000) Genetic analysis and conservation of 31 surviving individuals of a rare New Zealand tree, Metrosideros bartlettii (Myrtaceae). Molecular Ecology 9, 1149-1157. Dudley SA (1996) Differing selection on plant physiological traits in response to environmental water availability: A test of adaptive hypotheses. Evolution 50, 92-102. Dudley SA, Schmitt J (1996) Testing the adaptive plasticity hypothesis: Density-dependent selection on manipulated stem length in Impatiens capensis. American Naturalist 147, 445-465. 47 Dufresne F, Bourget E, Bernatchez L (2002) Differential patterns of spatial divergence in microsatellite and allozyme alleles: further evidence for locus-specific selection in the acorn barnacle, Semibalanus balanoides? Molecular Ecology 11, 113-123. Dutech C, Maggia L, Joly HI (2000) Chloroplast diversity in Vouacapoua americana (Caesalpiniaceae), a neotropical forest tree. Molecular Ecology 9, 14271432. Dutech C, Seiter J, Petronelli P, Joly HI, Jarne P (2002) Evidence of low gene flow in a neotropical clustered tree species in two rainforest stands of French Guiana. Molecular Ecology 11, 725-738. Eber S, Brandl R (1997) Genetic differentiation of the tephritid fly Urophora cardui in Europe as evidence for its biogeographical history. Molecular Ecology 6, 651-660. Echt CS, DeVerno LL, Anzidei M, Vendramin GG (1998) Chloroplast microsatellites reveal population genetic diversity in red pine, Pinus resinosa Ait. Molecular Ecology 7, 307-316. Ehinger M, Fontanillas P, Petit E, Perrin N (2002) Mitochondrial DNA variation along an altitudinal gradient in the greater white-toothed shrew, Crocidura russula. Molecular Ecology 11, 939-945. Einum S, Fleming IA (2000) Highly fecund mothers sacrifice offspring survival to maximize fitness. Nature 405, 565-567. Eizirik E, Kim JH, Menotti-Raymond M, et al. (2001) Phylogeography, population history and conservation genetics of jaguars (Panthera onca, Mammalia, Felidae). Molecular Ecology 10, 65-79. Eldridge MDB, Kinnear JE, Onus ML (2001) Source population of dispersing rock-wallabies (Petrogale lateralis) identified by assignment tests on multilocus genotypic data. Molecular Ecology 10, 2867-2876. ElMousadik A, Petit RJ (1996) Chloroplast DNA phylogeography of the argan tree of Morocco. Molecular Ecology 5, 547-555. Emerson BC, Wallis GB (1994) Species status and population genetic structure of the flightless chafer beetles Prodontria modesta and P. bicolorata (Coleoptera; Scarabaeidae) from South Island, New Zealand. Molecular Ecology 3, 339-345. Encalada SE, Lahanas PN, Bjorndal KA, et al. (1996) Phylogeography and population structure of the Atlantic and Mediterranean green turtle Chelonia mydas: a mitochondrial control region sequence assessment. Molecular Ecology 5, 473-483. England PR, Usher AV, Whelan RJ, Ayre DJ (2002) Microsatellite diversity and genetic structure of fragmented populations of the rare, fire-dependent shrub Grevillea macleayana. Molecular Ecology 11, 967-977. Escorza-Trevino S, Dizon AE (2000) Phylogeography, intraspecific structure and sex-biased dispersal of Dall's porpoise, Phocoenoides dalli, revealed by mitochondrial and microsatellite DNA analyses. Molecular Ecology 9, 1049-1060. Estoup A, Solignace M, Cornuet J-M, Goudet J, Scholl A (1996) Genetic differentiation of continental and island populations of Bombus terrestris (Hymenoptera: Aplidae) in Europe. Molecular Ecology 5, 19-31. Estoup S, Rousset R, Michalakis Y, et al. (1998) Comparative analysis of microsatellite and allozyme markers: a case study investigating microgeographic differentiation in brown trout (Salmo trutta). Molecular Ecology 7, 339-353. Falahati-Rastegar M, Manners JG, Smartt J (1981) Effects of temperature and inoculum density on competition between races of Puccinia hordei. Transactions of the British Mycological Society 77, 359-368. Faulkes GG, Abbott DB, O'Brien HP, et al. (1997) Micro- and macrogeographical genetic structure of colonies of naked mole rats Heterocephalus glaber. Molecular Ecology 6, 615-628. Feldheim KA, Gruber SH, Ashley MV (2001) Population genetic structure of the lemon shark (Negaprion brevirostris) in the western Atlantic: DNA microsatellite variation. Molecular Ecology 10, 295-303. 48 Finch MO, Lambert DM (1996) Kinship and genetic divergence among populations of tuatara Sphenodon punctatus as revealed by minisatellite DNA profiling. Molecular Ecology 5, 651-658. Fineschi S, Taurchini D, Villani F, Vendramin GG (2000) Chloroplast DNA polymorphism reveals little geographical structure in Castanea sativa Mill. (Fagaceae) throughout southern European countries. Molecular Ecology 9, 1495-1503. Finston T (2002) Geographic patterns of population genetic structure in Mytilocypris (Ostracoda : Cyprididae): interpreting breeding systems, gene flow and history in species with differing distributions. Molecular Ecology 11, 1931-1946. Firestone KB, Elphinstone MS, Sherwin WB, Houldin BA (1999) Phylogeographical population structure of tiger quolls Dasyurus maculatus (Dasyuridae: Marsupialia), an endangered carnivorous marsupial. Molecular Ecology 8, 1613-1625. Fonseca DM, LaPointe DA, Fleischer RC (2000) Bottlenecks and multiple introductions: population genetics of the vector of avian malaria in Hawaii. Molecular Ecology 9, 1803-1814. Forcioli D, Saumitou-Laprade P, Valero M, Vernet P, Cuguen J (1998) Distribution of chloroplast DNA diversity within and among populations in gynodioecious Beta vulgaris ssp. maritima (Chenopodiaceae). Molecular Ecology 7, 1193-1204. Ford MJ (2000) Effects of natural selection on patterns of DNA sequence variation at the transferrin, somatolactin, and p53 genes within and among chinook salmon (Oncorhynchus tshawytscha) populations. Molecular Ecology 9, 843-855. Fowler EV, Houlden BA, Hoeben P, Timms P (2000) Genetic diversity and gene flow among southeastern Queensland koalas (Phascolarctos cinereus). Molecular Ecology 9, 155-164. Freville H, Justy F, Olivieri I (2001) Comparative allozyme and microsatellite population structure in a narrow endemic plant species, Centaurea corymbosa Pourret (Asteraceae). Molecular Ecology 10, 879-889. Friar EA, Boose DL, LaDoux T, Roalson EH, Robichaux RH (2001) Population structure in the endangered Mauna Loa silversword, Argyroxiphium kauense (Asteraceae), and its bearing on reintroduction. Molecular Ecology 10, 1657-1663. Friesen VL, Congdon BC, Walsh HE, Birt TP (1997) Intron variation in marbled murrelets detected using analyses of single-stranded conformational polymorphisms. Molecular Ecology 6, 1047-1058. Friesen VL, Montevecchi WA, Baker AJ, Barrett RT, Davidson WS (1996) Population differentation and evolution in the common guillemot Uria aalge. Molecular Ecology 5, 793-805. Fry AJ, Zink RM (1998) Geographic analysis of nucleotide diversity and song sparrow (Aves: Emberizidae) population history. Molecular Ecology 7, 13031313. Frydenberg J, Pertoldi C, Dahlgaard J, Loeschcke V (2002) Genetic variation in original and colonizing Drosophila buzzatii populations analysed by microsatellite loci isolated with a new PCR screening method. Molecular Ecology 11, 181-190. Fullard KJ, Early G, Heide-Jorgensen MP, et al. (2000) Population structure of long-finned pilot whales in the North Atlantic: a correlation with sea surface temperature? Molecular Ecology 9, 949-958. Fuller SJ, Wilson JC, Mather PB (1997) Patterns of differentiation among wild rabbit populations Oryctolagus cuniculus L. in arid and semiarid ecosystems of north-eastern Australia. Molecular Ecology 6, 145-153. Fumagalli L, Moritz C, Taberlet P, Friend JA (1999) Mitochondrial DNA sequence variation within the remnant populations of the endangered numbat (Marsupialia : Myrmecobiidae : Myrmecobius fasciatus). Molecular Ecology 8, 1545-1549. Fumagalli L, Snoj A, Jesenek D, et al. (2002) Extreme genetic differentiation among the remnant populations of marble trout (Salmo marmoratus) in Slovenia. Molecular Ecology 11, 2711-2716. 49 Gabrielsen TM, Bachmann K, Jakobsen KS, Brochmann C (1997) Glacial survival does not matter: RAPD phylogeography of Nordic Saxifraga oppositifolia. Molecular Ecology 6, 831-842. Gabrielsen TM, Brochmann C (1998) Sex after all: high levels of diversity detected in the arctic clonal plant Saxifraga cernua using RAPD markers. Molecular Ecology 7, 1701-1708. Gabrielsen TM, Brochmann C, Rueness J (2002) The Baltic Sea as a model system for studying postglacial colonization and ecological differentiation, exemplified by the red alga Ceramium tenuicorne. Molecular Ecology 11, 2083-2095. Gadau GL, Heinze J, Hölldobler B, Schmid M (1996) Population and colony structure of the carpenter ant Camponotus floridanus. Molecular Ecology 5, 785792. Garant D, Dodson JJ, Bernatchez L (2000) Ecological determinants and temporal stability of the within- river population structure in Atlantic salmon (Salmo salar L.). Molecular Ecology 9, 615-628. García de León FJ, Chikhi L, Bonhomme F (1997) Microsatellite polymorphism and population subdivision in natural populations of European sea bass Dicentrarchus labrax (Linnaeus, 1758). Molecular Ecology 6, 51-62. Garcia RA, Zapater JMM, Criado BG, Zabalgogeazcoa I (2002) Genetic structure of natural populations of the grass endophyte Epichloe festucae in semiarid grasslands. Molecular Ecology 11, 355-364. Garcia-Martinez J, Moya A, Raga JA, Latorre A (1999) Genetic differentiation in the striped dolphin Stenella coeruleoalba from European waters according to mitochondrial DNA (mtDNA) restriction analysis. Molecular Ecology 8, 1069-1073. Garcia-Rodriguez AI, Bowen BW, Domning D, et al. (1998) Phylogeography of the West Indian manatee (Trichechus manatus): how many populations and how many taxa? Molecular Ecology 7, 1137-1149. Gaudeul M, Taberlet P, Till-Bottraud I (2000) Genetic diversity in an endangered alpine plant, Eryngium alpinum L.. (Apiaceae), inferred from amplified fragment length polymorphism markers. Molecular Ecology 9, 1625-1637. Ge XJ, Sun M (1999) Reproductive biology and genetic diversity of a cryptoviviparous mangrove Aegiceras corniculatum (Myrsinaceae) using allozyme and intersimple sequence repeat (ISSR) analysis. Molecular Ecology 8, 2061-2069. Gerlach G, Hoeck HN (2001) Islands on the plains: metapopulation dynamics and female biased dispersal in hyraxes (Hyracoidea) in the Serengeti National Park. Molecular Ecology 10, 2307-2317. Gibbs HL, Dawson RJG, Hobson KA (2000) Limited differentiation in microsatellite DNA variation among northern populations of the yellow warbler: evidence for male- biased gene flow? Molecular Ecology 9, 2137-2147. Gibbs HL, Prior KA, Weatherhead PJ, Johnson G (1997) Genetic structure of populations of the threatened eastern massasauga rattlesnake, Sistrurus c. catenatus: evidence from microsatellite DNA markers. Molecular Ecology 6, 1123-1132. Gillies ACM, Cornelius JP, Newton AC, et al. (1997) Genetic variation in Costa Rican populations of the tropical timber species Cedrela odorata L., assessed using RAPDs. Molecular Ecology 6, 1133-1145. Giraud T, Blatrix R, Poteaux C, Solignac M, Jaisson P (2000) Population structure and mating biology of the polygynous ponerine ant Gnamptogenys striatula in Brazil. Molecular Ecology 9, 1835-1841. Girman D, Vila C, Geffen E, et al. (2001) Patterns of population subdivision, gene flow and genetic variability in the African wild dog (Lycaon pictus). Molecular Ecology 10, 1703-1723. Gleason JM, Nuzhdin SV, Ritchie MG (2002) Quantitative trait loci affecting a courtship signal in Drosophila melanogaster. Heredity 89, 1-6. Godt MJW, Hamrick JL (1999) Population genetic analysis of Elliottia racemosa (Ericaceae), a rare Georgia shrub. Molecular Ecology 8, 75-82. 50 Gomes C, Dales RBG, Oxenford HA (1998) The application of RAPD markers in stock discrimination of the four-wing flying fish, Hirunichthys affinis in the central western Atlantic. Molecular Ecology 7, 1029-1039. Gomulski LM, Bourtzis K, Brogna S, et al. (1998) Intron size polymorphism of the Adh1 gene parallels the worldside colonization history of the Mediterranean fruit fly, Ceratitis capitata. Molecular Ecology 7, 1729-1741. González S, Maldonado JE, Leonard JA, et al. (1998) Conservation genetics of the endangered Pampas deer (Ozotoceros bezoarticus). Molecular Ecology 7, 4756. Goodacre SL (2002) Population structure, history and gene flow in a group of closely related land snails: genetic variation in Partula from the Society Islands of the Pacific. Molecular Ecology 11, 55-68. Goodisman MAD, Matthews RW, Crozier RH (2001) Hierarchical genetic structure of the introduced wasp Vespula germanica in Australia. Molecular Ecology 10, 1423-1432. Goodman SJ, Tamate HB, Wilson R, et al. (2001) Bottlenecks, drift and differentiation: the population structure and demographic history of sika deer (Cervus nippon) in the Japanese archipelago. Molecular Ecology 10, 1357-1370. Goostrey S, Carss DN, Noble LR, Piertney SB (1998) Population introgression and differentiation in the great cormorant Phalacrocorax carbo in Europe. Molecular Ecology 7, 329-338. Gordon TR, Okamoto D, Milgroom MG (1992) The structure and interrelationship of fungal populations in native and cultivated soils. Molecular Ecology 1, 231-249. Goropashnaya AV, Seppa P, Pamilo P (2001) Social and genetic characteristics of geographically isolated populations in the ant Formica cinerea. Molecular Ecology 10, 2807-2818. Gosselin L, Jobidon R, Bernier L (1999) Genetic variability and structure of Canadian populations of Chondrostereum purpureum, a potential biophytocide. Molecular Ecology 8, 113-122. Gottelli D, Sillero-Zubiri C, Applebaum GD, et al. (1994) Molecular genetics of the most endangered canid: the Ethiopian wolf Canis simensis. Molecular Ecology 3, 301-312. Grapputo A, Pilastro A, Marin G (1998) Genetic variation and bill size dimorphism in a passerine bird. Molecular Ecology 7, 1173-1182. Grashof-Bokdam CJ, Jansen J, Smulders MJM (1998) Dispersal patterns of Lonicera periclymenum determined by genetic analysis. Molecular Ecology 7, 165174. Green JM, Barker JHA, Marshall EJP, et al. (2001) Microsatellite analysis of the inbreeding grass weed Barren Brome (Anisantha sterilis) reveals genetic diversity at the within- and between-farm scales. Molecular Ecology 10, 1035-1045. Grunwald C, Stabile J, Waldman JR, Gross R, Wirgin I (2002) Population genetics of shortnose sturgeon Acipenser brevirostrum based on mitochondrial DNA control region sequences. Molecular Ecology 11, 1885-1898. Gugerli F, Eichenberger K, Schneller JJ (1999) Promiscuity in populations of the cushion plant Saxifraga oppositifolia in the Swiss Alps as inferred from random amplified polymorphic DNA (RAPD). Molecular Ecology 8, 453-461. Gugerli F, Sperisen C, Buchler U, et al. (2001) Haplotype variation in a mitochondrial tandem repeat of Norway spruce (Picea abies) populations suggests a serious founder effect during postglacial re-colonization of the western Alps. Molecular Ecology 10, 1255-1263. Gurganus MC, Fry JD, Nuzhdin SV, et al. (1998) Genotype-environment interaction at quantitative trait loci affecting sensory bristle number in Drosophila melanogaster. Genetics 149, 1883-1898. Gustafsson S (2000) Patterns of genetic variation in Gymnadenia conopsea, the fragrant orchid. Molecular Ecology 9, 1863-1872. 51 Haavie J, Saetre GP, Moum T (2000) Discrepancies in population differentiation at microsatellites, mitochondrial DNA and plumage colour in the pied flycatcher - inferring evolutionary processes. Molecular Ecology 9, 1137-1148. Hagen MJ, Hamrick JL (1996) A hierarchical analysis of population genetic structure in Rhizobium leguminosarum bv. Trifolii. Molecular Ecology 5, 177-186. Haig SM, Bowman B, Mullins TD (1996) Population structure of red-cockaded woodpeckers in south Florida: RAPDs revisited. Molecular Ecology 5, 725-734. Haig SM, Gratto-Trevor CL, Mullins TD, Colwell MA (1997) Population identification of western hemisphere songbirds throughout the annual cycle. Molecular Ecology 6, 413-427. Haig SM, Rhymer JM, Heckel DG (1994) Population differentation in randomly amplified polymorphic DNA of red-cockaded woodpeckers Picoides borealis. Molecular Ecology 3, 581-595. Hänfling B, Brandl R (1998) Genetic variability, population size and isolation of distinct populations in the freshwater fish Cottus gobio L. Molecular Ecology 7, 1625-1632. Hanfling B, Hellemans B, Volckaert FAM, Carvalho GR (2002) Late glacial history of the cold-adapted freshwater fish Cottus gobio, revealed by microsatellites. Molecular Ecology 11, 1717-1729. Hansen MM, Mensberg KLD, Berg S (1999) Postglacial recolonization patterns and genetic relationships among whitefish (Coregonus sp.) populations in Denmark, inferred from mitochondrial DNA and microsatellite markers. Molecular Ecology 8, 239-252. Hansen MM, Ruzzante DE, Nielsen EE, Bekkevold D, Mensberg K-LD (2002) Long-term effective population sizes, temporal stability of genetic composition and potential for local adaptation in anadromous brown trout (Salmo trutta) populations. Molecular Ecology 11, 2523-2535. Hardy ME, Grady JM, Routman EJ (2002) Intraspecific phylogeography of the slender madtom: the complex evolutionary history of the Central Highlands of the United States. Molecular Ecology 11, 2393-2403. Hauser L, Carvalho GR, Pitcher TJ, Ogutu-Ohwayo R (1998) Genetic affinities of an introduced predator: Nile perch in Lake Victoria, East Africa. Molecular Ecology 7, 849-857. Hawdon JM, Li T, Zhan B, Blouin MS (2001) Genetic structure of populations of the human hookworm, Necator americanus, in China. Molecular Ecology 10, 1433-1437. Hawthorne DJ, Via S (2001) Genetic linkage of ecological specialization and reproductive isolation in pea aphids. Nature 412, 904-907. He M, Haymer DS (1999) Genetic relationships of populations and the origins of new infestations of the Mediterranean fruit fly. Molecular Ecology 8, 12471257. Hebert C, Danzman RG, Jones MW, Bernatchez L (2000) Hydrography and population genetic structure in brook charr (Salvelinus fontinalis, Mitchill) from eastern Canada. Molecular Ecology 9, 971-982. Heuertz M, Hausman JF, Tsvetkov I, Frascaria-Lacoste N, Vekemans X (2001) Assessment of genetic structure within and among Bulgarian populations of the common ash (Fraxinus excelsior L.). Molecular Ecology 10, 1615-1623. Hoarau G, Rijnsdorp AD, Van der Veer HW, Stam WT, Olsen JL (2002) Population structure of plaice (Pleuronectes platessa L.) in northern Europe: microsatellites revealed large-scale spatial and temporal homogeneity. Molecular Ecology 11, 1165-1176. Högberg N, J. Stenlid, Karlsson J-O (1995) Genetic differentiation in Fomitopsis pinicola (Schwarts: Fr.) Karst studied by means of arbitrary-primed PCR. Molecular Ecology 4, 675-680. Hogberg N, Stenlid J (1999) Population genetics of Fomitopsis rosea - a wood-decay fungus of the old-growth European taiga. Molecular Ecology 8, 703-710. Holder K, Montgomerie R, Friesen VL (2000) Glacial vicariance and historical biogeography of rock ptarmigan (Lagopus mutus) in the Bering region. Molecular Ecology 9, 1265-1278. 52 Holderegger R, Stehlik I, Abbott RJ (2002) Molecular analysis of the Pleistocene history of Saxifraga oppositifolia in the Alps. Molecular Ecology 11, 14091418. Holland BS, Hadfield MG (2002) Islands within an island: phylogeography and conservation genetics of the endangered Hawaiian tree snail Achatinella mustelina. Molecular Ecology 11, 365-375. Houlden BA, Costello BH, Sharkey D, et al. (1999) Phylogeographic differentiation in the mitochondrial control region in the koala, Phascolarctos cinereus (Goldfuss 1817). Molecular Ecology 8, 999-1011. Houlden BA, England PR, Taylor AC, Greville WD, Sherwin WB (1996) Low genetic variability of the koala Phascolarctos cinereus in south-eastern Australia following a severe population bottleneck. Molecular Ecology 5, 269-281. Hsiao JY, Lee SM (1999) Genetic diversity and microgeographic differentiation of Yushan cane (Yushania niitakayamensis ; Poaceae) in Taiwan. Molecular Ecology 8, 263-270. Huang S, Chiang YC, Schaal BA, Chou CH, Chiang TY (2001) Organelle DNA phylogeography of Cycas taitungensis, a relict species in Taiwan. Molecular Ecology 10, 2669-2681. Huang SSF, Hwang S-Y, Lin T-P (2002) Spatial pattern of chloroplast DNA variation of Cyclobalanopsis glauca in Taiwan and East Asia. Molecular Ecology 11, 2349-2358. Huber K, Le Loan L, Hoang TH, et al. (2002) Genetic differentiation of the dengue vector, Aedes aegypti (Ho Chi Minh City, Vietnam) using microsatellite markers. Molecular Ecology 11, 1629-1635. Huff DR, Quinn JA, Higgins B, Palazzo AJ (1998) Random amplified polymorphic DNA (RAPD) variation among native little bluestem Schizachyrium scoparium (Michx.) Nash populations from sites of high and low fertility in forest and grassland biomes. Molecular Ecology 7, 1591-1597. Hurwood DA, Hughes JM (1998) Phylogeography of the freshwater fish, Mogurnda adspersa, in streams of northeastern Queensland, Australia: evidence for altered drainage patterns. Molecular Ecology 7, 1507-1517. James CH, Moritz C (2000) Intraspecific phylogeography in the sedge frog Litoria fallax (Hylidae) indicates pre-Pleistocene vicariance of an open forest species from eastern Australia. Molecular Ecology 9, 349-358. James TY, Vilgalys R (2001) Abundance and diversity of Schizophyllum commune spore clouds in the Caribbean detected by selective sampling. Molecular Ecology 10, 471-479. Janzen FJ (1993) An Experimental-Analysis of Natural-Selection on Body Size of Hatchling Turtles. Ecology 74, 332-341. Janzen FJ (1995) Experimental-Evidence for the Evolutionary Significance of Temperature-Dependent Sex Determination. Evolution 49, 864-873. Jaramillo-Correa JP, Beaulieu J, Bousquet J (2001) Contrasting evolutionary forces driving population structure at expressed sequence tag polymorphisms, allozymes and quantitative traits in white spruce. Molecular Ecology 10, 2729-2740. Jehle R, Arntzen W, Burke T, Krupa AP, Hodl W (2001) The annual number of breeding adults and the effective population size of syntopic newts (Triturus cristatus, T- marmoratus). Molecular Ecology 10, 839-850. Jerome CA, Ford BA (2002) The discovery of three genetic races of the dwarf mistletoe Arceuthobium americanum (Viscaceae) provides insight into the evolution of parasitic angiosperms. Molecular Ecology 11, 387-405. Jerry DR, Baverstock PR (1998) Consequences of a catadromous life-strategy for levels of mitochondrial DNA differentiation among populations of the Australian bass, Macquaria novemaculeata. Molecular Ecology 7, 1003-1013. Johannesen J, Veith M, Seitz A (1996) Population genetic structure of the butterfly Melitaea didyma (Nymphalidae) along a northern distribution border. Molecular Ecology 5, 259-267. 53 Johannesson H, Vasiliauskas R, Dahlberg A, Penttila R, Stenlid J (2001) Genetic differentiation in Eurasian populations of the postfire ascomycete Daldinia loculata. Molecular Ecology 10, 1665-1677. Jones CD (1998) The genetic basis of Drosophila sechellia's resistance to a host plant toxin. Genetics 149, 1899-1908. Jones CD (2001) The genetic basis of larval resistance to a host plant toxin in Drosophila sechellia. Genetical Research 78, 225-233. Jordano P, Godoy JA (2000) RAPD variation and population genetic structure in Prunus mahaleb (Rosaceae), an animal-dispersed tree. Molecular Ecology 9, 1293-1305. Katsuya K, Green GJ (1967) Reproductive potentials of races 15B and 56 of wheat stem rust. Canadian Journal of Botany-Revue Canadienne De Botanique 45, 1077-1091. Keightley PD, Hardge T, May L, Bulfield G (1996) A genetic map of quantitative trait loci for body weight in the mouse. Genetics 142, 227-235. Keiper FJ, McConchie R (2000) An analysis of genetic variation in natural populations of Sticherus flabellatus R. Br. (St John) using amplified fragment length polymorphism (AFLP) markers. Molecular Ecology 9, 571-581. Kerth G, Mayer F, Konig B (2000) Mitochondrial DNA (mtDNA) reveals that female Bechstein's bats live in closed societies. Molecular Ecology 9, 793-800. Kerth G, Mayer F, Petit E (2002) Extreme sex-biased dispersal in the communally breeding, nonmigratory Bechstein's bat (Myotis bechsteinii). Molecular Ecology 11, 1491-1498. Kim I, Phillips CJ, Monjeau JA, et al. (1998) Habitat islands, genetic diversity, and gene flow in a Patagonian rodent. Molecular Ecology 7. Kimura M, Clegg SM, Lovette IJ, et al. (2002) Phylogeographical approaches to assessing demographic connectivity between breeding and overwintering regions in a Nearctic-Neotropical warbler (Wilsonia pusilla). Molecular Ecology 11, 1605-1616. King RA, Gornall RJ, Preston CD, Croft JM (2002) Population differentiation of Potamogeton pectinatus in the Baltic Sea with reference to waterfowl dispersal. Molecular Ecology 11, 1947-1956. Klingenberg CP, Leamy LJ, Routman EJ, Cheverud JM (2001) Genetic architecture of mandible shape in mice: Effects of quantitative trait loci analyzed by geometric morphometrics. Genetics 157, 785-802. Knight CG, Azevedo RBR, Leroi AM (2001) Testing life-history pleiotropy in Caenorhabditis elegans. Evolution 55, 1795-1804. Knowles LL (2001) Did the Pleistocene glaciations promote divergence? Tests of explicit refugial models in montane grasshopprers. Molecular Ecology 10, 691701. Kolliker R, Stadelmann FJ, Reidy B, Nosberger J (1998) Fertilization and defoliation frequency affect genetic diversity of Festuca pratensis Huds. in permanent grasslands. Molecular Ecology 7, 1557-1567. Kontula T, Vainola R (2001) Postglacial colonization of Northern Europe by distinct phylogeographic lineages of the bullhead, Cottus gobio. Molecular Ecology 10, 1983-2002. Koskinen MT, Knizhin I, Primmer CR, Schlötterer C, Weiss S (2002) Mitochondrial and nuclear DNA phylogeography of Thymallus spp. (grayling) provides evidence of ice-age mediated environmental perturbations in the world's oldest body of fresh water, Lake Baikal. Molecular Ecology 11, 2599-2611. Koskinen MT, Ranta E, Piironen J, et al. (2000) Genetic lineages and postglacial colonization of grayling (Thymallus thymallus, Salmonidae) in Europe, as revealed by mitochondrial DNA analyses. Molecular Ecology 9, 1609-1624. Krieger MJB, Keller L (2000) Mating frequency and genetic structure of the Argentine ant Linepithema humile. Molecular Ecology 9, 119-126. Kropf M, Kadereit JW, Comes HP (2002) Late Quaternary distributional stasis in the submediterranean mountain plant Anthyllis montana L. (Fabaceae) inferred from ITS sequences and amplified fragment length polymorphism markers. Molecular Ecology 11, 447-463. 54 Kruger AM, Hellwig FH, Oberprieler C (2002) Genetic diversity in natural and anthropogenic inland populations of salt-tolerant plants: random amplified polymorphic DNA analyses of Aster tripolium L. (Compositae) and Salicornia ramosissima Woods (Chenopodiaceae). Molecular Ecology 11, 16471655. Kumar A, Rogstad SH (1998) A hierarchical analysis of minisatellite DNA diversity in Gambel oak (Quercus gambelii Nutt.; Fagaceae). Molecular Ecology 7, 859-869. Kwon JA, Morden CW (2002) Population genetic structure of two rare tree species (Colubrina oppositifolia and Alphitonia ponderosa, Rhamnaceae) from Hawaiian dry and mesic forests using random amplified polymorphic DNA markers. Molecular Ecology 11, 991-1001. Kyle CJ, Robitaille JF, Strobeck C (2001) Genetic variation and structure of fisher (Martes pennanti) populations across North America. Molecular Ecology 10, 2341-2347. Kyle CJ, Strobeck C (2001) Genetic structure of North American wolverine (Gulo gulo) populations. Molecular Ecology 10, 337-347. Lacerda DR, Acedo MDP, Lemos JP, Lovato B (2001) Genetic diversity and structure of natural populations of Plathymenia reticulata (Mimosoideae), a tropical tree from the Brazilian Cerrado. Molecular Ecology 10, 1143-1152. Lade JA, Murray ND, Marks CA, Robinson NA (1996) Microsatellite differentiation between Phillip Island and mainland Australian populations of the red fox Vulpes vulpes. Molecular Ecology 5, 81-87. Ladoukakis ED, Saavedra C, Magoulas A, Zouros E (2002) Mitochondrial DNA variation in a species with two mitochondrial genomes: the case of Mytilus galloprovincialis from the Atlantic, the Mediterranean and the Black Sea. Molecular Ecology 11, 755-769. Largiader CR, Herger F, Lortscher M, Scholl A (2000) Assessment of natural and artificial propagation of the white- clawed crayfish (Austropotamobius pallipes species complex) in the Alpine region with nuclear and mitochondrial markers. Molecular Ecology 9, 25-37. Laurent L, Casale P, Bradai MN, et al. (1998) Molecular resolution of marine turtle stock composition in fishery bycatch: a case study in the Mediterranean. Molecular Ecology 7, 1529-1542. Lavery S, Moritz C, Fielder DR (1996) Indo-Pacific population structure and evolutionary history of the coconut crab Birgus latro. Molecular Ecology 5, 557570. Le Roy I, Tordjman S, Migliore-Samour D, Degrelle H, Roubertoux PL (2001) Genetic architecture of testis and seminal vesicle weights in mice. Genetics 158, 333-340. LeCorre V, DumolinLapegue S, Kremer A (1997) Genetic variation at allozyme and RAPD loci in sessile oak Quercus petraea (Matt) Liebl: The role of history and geography. Molecular Ecology 6, 519-529. Lee PLM, Bradbury RB, Wilson JD, et al. (2001) Microsatellite variation in the yellowhammer Emberiza citrinella: population structure of a declining farmland bird. Molecular Ecology 10, 1633-1644. Lehmann T, Besansky NJ, Hawley WA, et al. (1997) Microgeographic structure of Anopheles gambiae in western Kenya based on mtDNA and microsatellite loci. Molecular Ecology 6, 243-253. Leips J, Mackay TFC (2000) Quantitative trait loci for life span in Drosophila melanogaster: Interactions with genetic background and larval density. Genetics 155, 1773-1788. Lemaire C, Allegrucci G, Naciri M, et al. (2000) Do discrepancies between microsatellite and allozyme variation reveal differential selection between sea and lagoon in the sea bass (Dicentrarchus labrax)? Molecular Ecology 9, 457-467. Lenk P, Fritz U, Joger U, Winks M (1999) Mitochondrial phylogeography of the European pond turtle, Emys orbicularis (Linnaeus 1758). Molecular Ecology 8, 1911-1922. Leonard KJ (1969) Selection in heterogeneous populations of Puccinia graminis f.sp. avenae. Phytopathology 59, 1851-1857. 55 Leonard KJ (1977) Virulence, temperature optima and competitive abilities of isolines of races T and O of Bipolar maydis. Phytopathology 67, 1273-1279. Lepetit D, Brehm A, Fouillet P, Biemont C (2002) Insertion polymorphism of retrotransposable elements in populations of the insular, endemic species Drosophila madeirensis. Molecular Ecology 11, 347-354. Liao LC, Hsiao JY (1998) Relationship between population genetic structure and riparian habitat as revealed by RAPD analysis of the rheophyte Acorus gramineus Soland. (Araceae) in Taiwan. Molecular Ecology 7, 1275-1281. Liu J, Mercer JM, Stam LF, et al. (1996) Genetic analysis of a morphological shape difference in the male genitalia of Drosophila simulans and D-mauritiana. Genetics 142, 1129-1145. Llopart A, Elwyn S, Lachaise D, Coyne JA (2002) GENETICS OF A DIFFERENCE IN PIGMENTATION BETWEEN DROSOPHILA YAKUBA AND DROSOPHILA SANTOMEA. Evolution 56, 2262-2277. Loegering WQ (1951) Survival of races of wheat stem rust in mixtures. Phytopathology 41, 56-65. Lugon-Moulin N, Brunner H, Wyttenbach A, Hausser J, Goudet J (1999) Hierarchical analyses of genetic differentiation in a hybrid zone of Sorex araneus (Insectivora : Soricidae). Molecular Ecology 8, 419-431. Lugon-Moulin N, Hausser J (2002) Phylogeographical structure, postglacial recolonization and barriers to gene flow in the distinctive Valais chromosome race of the common shrew (Sorex araneus). Molecular Ecology 11, 785-794. Lundy CJ, Moran P, Rico C, Milner RS, Hewitt GM (1999) Macrogeographical population differentiation in oceanic environments: a case study of European Hake (Merluccius merluccius), a commercially important fish. Molecular Ecology 8, 1889-1898. Lundy CJ, Rico C, Hewitt GM (2000) Temporal and spatial genetic variation in spawning grounds of European hake (Merluccius merluccius) in the Bay of Biscay. Molecular Ecology 9, 2067-2079. MacDonald C, Brookfield JFY (2002) Intraspecific molecular variation in the seaweed fly Coelopa frigida consistent with behavioural distinctness of British and Swedish populations. Molecular Ecology 11, 1637-1646. Macdonald SJ, Goldstein DB (1999) A quantitative genetic analysis of male sexual traits distinguishing the sibling species Drosophila simulans and D. sechellia. Genetics 153, 1683-1699. Machordom A, Suarez J, Almodovar A, Bautista JM (2000) Mitochondrial haplotype variation and phylogeography of Iberian brown trout populations. Molecular Ecology 9, 1325-1338. Maguire TL, Saenger P, Baverstock P, Henry R (2000) Microsatellite analysis of genetic structure in the mangrove species Avicennia marina (Forsk.) Vierh. (Avicenniaceae). Molecular Ecology 9, 1853-1862. Maki M, Horie S (1999) Random amplified polymorphic DNA (RAPD) markers reveal less genetic variation in the endangered plant Cerastium fischerianum var. molle than in the widespread conspecific C- fischerianum var. fischerianum (Caryophyllaceae). Molecular Ecology 8, 145-150. Maldonado JE, Vila C, Wayne RK (2001) Tripartite genetic subdivisions in the ornate shrew (Sorex ornatus). Molecular Ecology 10, 127-147. Markert JA, Arnegard ME, Danley PD, Kocher TD (1999) Biogeography and population genetics of the Lake Malawi cichlid Melanochromis auratus: habitat transience, philopatry and speciation. Molecular Ecology 8, 1013-1026. Marshall HD, Ritland K (2002) Genetic diversity and differentiation of Kermode bear populations. Molecular Ecology 11, 685-697. Martens JW (1973) Competitive ability of oat stem rust races in mixtures. Canadian Journal of Botany-Revue Canadienne De Botanique 51, 2233-2236. Martin C, GonzalezBenito ME, Iriondo JM (1997) Genetic diversity within and among populations of a threatened species: Erodium paularense Fern Gent & Izco. Molecular Ecology 6, 813-820. Martin C, Gonzalez-Benito ME, Iriondo JM (1999) The use of genetic markers in the identification and characterization of three recently discovered populations of a threatened plant species. Molecular Ecology 8, S31-S40. 56 Martínez-Torres D, Simon JC, Fereres A, Moya A (1996) Genetic variation in natural populations of the aphid Rhopalosiphum padi as revealed by maternally inherited markers. Molecular Ecology, 659-670. Massonnet B, Simon J-C, Weisser WW (2002) Metapopulation structure of the specialized herbivore Macrosiphoniella tanacetaria (Homoptera, Aphididae). Molecular Ecology 11, 2511-2521. Mateu-Andres I, Segarra-Moragues JG (2000) Population subdivision and genetic diversity in two narrow endemics of Antirrhinum L. Molecular Ecology 9, 2081-2087. Maudet C, Miller C, Bassano B, et al. (2002) Microsatellite DNA and recent statistical methods in wildlife conservation management: applications in Alpine ibex Capra ibex (ibex). Molecular Ecology 11, 421-436. Mavarez J, Amarista M, Pointier JP, Jarne P (2002) Fine-scale population structure and dispersal in Biomphalaria glabrata, the intermediate snail host of Schistosoma mansoni, in Venezuela. Molecular Ecology 11, 879-889. McCartney MA, Keller G, Lessios HA (2000) Dispersal barriers in tropical oceans and speciation in Atlantic and eastern Pacific sea urchins of the genus Echinometra. Molecular Ecology 9, 1391-1400. McConnell SKJ, Ruzzante DE, O'Reilly PT, Hamilton L, Wright JM (1997) Microsatellite loci reveal highly significant genetic differentiation among Atlantic salmon (Salmo salar L.) stocks from the east coast of Canada. Molecular Ecology 6, 1075-1089. McGee DC, Zuck MG (1981) Competition between benomyl-resistant and sensitive strains of Venturia inaequalis on apple seedlings. Phytopathology 71, 529532. McGlashan DJ, Hughes JM (2000) Reconciling patterns of genetic variation with stream structure, earth history and biology in the Australian freshwater fish Craterocephalus stercusmuscarum (Atherinidae). Molecular Ecology 9, 1737-1751. McGregor R (1998) Evolution of life-history timing in a leafmining moth: Phenotypic selection in patches with manipulated development time. Evolutionary Ecology 12, 629-642. McGuigan K, McDonald K, Parris K, Moritz C (1998) Mitochondrial DNA diversity and historical biogeography of a wet forest-restricted frog (Litoria pearsoniana) from mid-east Australia. Molecular Ecology 7. McLean JE, Hay DE, Taylor EB (1999) Marine population structure in an anadromous fish: life-history influences patterns of mitochondrial DNA variation in the eulachon, Thaleichthys pacificus. Molecular Ecology 8, S143-S158. McMillan WO, Bermingham E (1996) The phylogeographic pattern of mitochondrial DNA variation in Dall's porpoise Phocoenoides dalli. Molecular Ecology 5, 47-61. Mengoni A, Barabesi C, Gonnelli C, et al. (2001) Genetic diversity of heavy metal-tolerant populations in Silene paradoxa L. (Caryophyllaceae): a chloroplast microsatellite analysis. Molecular Ecology 10, 1909-1916. Mengoni A, Gonnelli C, Galardi F, Gabbrielli R, Bazzicalupo M (2000) Genetic diversity and heavy metal tolerance in populations of Silene paradoxa L. (Caryophyllaceae): a random amplified polymorphic DNA analysis. Molecular Ecology 9, 1319-1324. Merila J, Przybylo R, Sheldon BC (1999) Genetic variation and natural selection on blue tit body condition in different environments. Genetical Research 73, 165-176. Meunier C, Tirard C, Hurtrez-Bousses S, et al. (2001) Lack of molluscan host diversity and the transmission of an emerging parasitic disease in Bolivia. Molecular Ecology 10, 1333-1340. Michels E, Cottenie K, Neys L, et al. (2001) Geographical and genetic distances among zooplankton populations in a set of interconnected ponds: a plea for using GIS modelling of the effective geographical distance. Molecular Ecology 10, 1929-1938. 57 Milot E, Gibbs HL, Hobson KA (2000) Phylogeography and genetic structure of northern populations of the yellow warbler (Dendroica petechia). Molecular Ecology 9, 667-681. Mitton JB, Kreiser BR, Latta RG (2000) Glacial refugia of limber pine (Pinus flexilis James) inferred from the population structure of mitochondrial DNA. Molecular Ecology 9, 91-97. Mock KE, Theimer TC, Rhodes OE, Greenberg DL, Keim P (2002) Genetic variation across the historical range of the wild turkey (Meleagris gallopavo). Molecular Ecology 11, 643-657. Mockford SW, Snyder M, Herman TB (1999) A preliminary examination of genetic variation in a peripheral population of Blanding's turtle, Emydoidea blandingii. Molecular Ecology 8, 323-327. Morand ME, Brachet S, Rossignol P, Dufour J, Frascaria-Lacoste N (2002) A generalized heterozygote deficiency assessed with microsatellites in French common ash populations. Molecular Ecology 11, 377-385. Morden CW, Loeffler W (1999) Fragmentation and genetic differentiation among subpopulations of the endangered Hawaiian mint Haplostachys haplostachya (Lamiaceae). Molecular Ecology 8, 617-625. Morgan-Richards M, Wolff K (1999) Genetic structure and differentiation of Plantago major reveals a pair of sympatric sister species. Molecular Ecology 8, 1027-1036. Moritz C, Heideman A, Geffen E, McRae P (1997) Genetic population structure of the Greater Bilby Macrotis lagotis, a marsupial in decline. Molecular Ecology 6, 925-936. Mossman CA, Waser PM (1999) Genetic detection of sex-biased dispersal. Molecular Ecology 8, 1063-1067. Moya A, Guirao P, Cifuentes D, Beitia F, Cenis JL (2001) Genetic diversity of Iberian populations of Bemisia tabaci (Hemiptera : Aleyrodidae) based on random amplified polymorphic DNA-polymerase chain reaction. Molecular Ecology 10, 891-897. Muller MH, Prosperi JM, Santoni S, Ronfort J (2001) How mitochondrial DNA diversity can help to understand the dynamics of wild-cultivated complexes. The case of Medicago sativa in Spain. Molecular Ecology 10, 2753-2763. Muller-Scharer H, Fischer M (2001) Genetic structure of the annual weed Senecio vulgaris in relation to habitat type and population size. Molecular Ecology 10, 17-28. Mundy NI, Winchell CS, Woodruff DS (1997) Genetic differences between the endangered San Clemente Island loggerhead shrike Lanius ludovicianus mearnsi and two neighbouring subspecies demonstrated by mtDNA control region and cytochrome b sequence variation. Molecular Ecology 6, 29-37. Murphy RW, Fu JZ, Upton DE, De Lema T, Zhao EM (2000) Genetic variability among endangered Chinese giant salamanders, Andrias davidianus. Molecular Ecology 9, 1539-1547. Nagata J, Masuda R, Kaji K, Kaneko M, Yoshida MC (1998) Genetic variation and population structure of the Japanese sika deer (Cervus nippon) in Hokkaido Island, based on mitochondrial D-loop sequences. Molecular Ecology 7, 871-878. Nersting LG, Arctander P (2001) Phylogeography and conservation of impala and greater kudu. Molecular Ecology 10, 711-719. Nesbo CL, Fossheim T, Vollestad LA, Jakobsen KS (1999) Genetic divergence and phylogeographic relationships among European perch (Perca fluviatilis) populations reflect glacial refugia and postglacial colonization. Molecular Ecology 8, 1387-1404. Newman RA, Squire T (2001) Microsatellite variation and fine-scale population structure in the wood frog (Rana sylvatica). Molecular Ecology 10, 1087-1100. Ng SC, Corlett RT (2000) Genetic variation and structure in six Rhododendron species (Ericaceae) with contrasting local distribution patterns in Hong Kong, China. Molecular Ecology 9, 959-969. Nielsen JL, Crow KD, Fountain MC (1999) Microsatellite diversity and conservation of a relic trout population: McCloud River redband trout. Molecular Ecology 8, S129-S142. 58 Nusser JA, Goto RM, Ledig DB, Fleischer RC, Miller MM (1996) RAPD analysis reveals low genetic variability in the endangered light-footed clapper rail. Molecular Ecology 5, 463-472. Nuzhdin SV, Pasyukova EG, Dilda CL, Zeng ZB, Mackay TFC (1997) Sex-specific quantitative trait loci affecting longevity in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 94, 9734-9739. Nuzhdin SV, Reiwitch SG (2000) Are the same genes responsible for intra- and interspecific variability for sex comb tooth number in Drosophila? Heredity 84, 97-102. Nyakaana S, Arctander P (1999) Population genetic structure of the African elephant in Uganda based on variation at mitochondrial and nuclear loci: evidence for male-biased gene flow. Molecular Ecology 8, 1105-1115. O'Corry-Crowe GM, Suydam RS, Rosenberg A, Frost KJ, Dizon AE (1997) Phylogeography, population structure and dispersal patterns of the beluga whale Delphinapterus leucas in the western Neartic revealed by mitochondrial DNA. Molecular Ecology 6, 955-970. Ohara RB, Brown JKM (1996) Frequency- and density-dependent selection in wheat powdery mildew. Heredity 77, 439-447. Onyabe DY, Conn JE (2001) Population genetic structure of the malaria mosquito Anopheles arabiensis across Nigeria suggests range expansion. Molecular Ecology 10, 2577-2591. Osoro MO, Green GJ (1976) Stabilizing selection in Puccinia graminis tritici in Canada. Canadian Journal of Botany-Revue Canadienne De Botanique 54, 22042214. Paetkau D, Amstrup SC, Born EW, et al. (1999) Genetic structure of the world's polar bear populations. Molecular Ecology 8, 1571-1584. Painter JN, Crozier RH, Poiani A, Robertson RJ, Clarke MF (2000) Complex social organization reflects genetic structure and relatedness in the cooperatively breeding bell miner, Manorina melanophrys. Molecular Ecology 9, 1339-1347. Palacios C, Gonzalez-Candelas F (1997) Analysis of population genetic structure and variability using RAPD markers in the endemic and endangered Limonium dufourii (Plumbaginaceae). Molecular Ecology 6, 1107-1121. Palacios C, Kresovich S, Gonzalez-Candelas F (1999) A population genetic study of the endangered plant species Limonium dufourii (Plumbaginaceae) based on amplified fragment length polymorphism (AFLP). Molecular Ecology 8, 645-657. Palme AE, Vendramin GG (2002) Chloroplast DNA variation, postglacial recolonization and hybridization in hazel, Corylus avellana. Molecular Ecology 11, 1769-1780. Palsson S (2000) Microsatellite variation in Daphnia pulex from both sides of the Baltic Sea. Molecular Ecology 9, 1075-1088. Parker MA (1994) Evolution in Natural and Experimental Populations of Amphicarpaea-Bracteata. Journal of Evolutionary Biology 7, 567-579. Patton JL, Silva MNFD, Malcolm JR (1996) Hierarchical genetic structure and gene flow in three sympatric species of Amazonian rodents. Molecular Ecology 5, 229-238. Peakall R, Smouse PE, Huff DR (1995) Evolutionary Implications of Allozyme and Rapd Variation in Diploid Populations of Dioecious Buffalograss Buchloe Dactyloides. Molecular Ecology 4, 135-147. Pearson CVM, Rogers AD, Sheader M (2002) The genetic structure of the rare lagoonal sea anemone, Nematostella vectensis Stephenson (Cnidaria; Anthozoa) in the United Kingdom based on RAPD analysis. Molecular Ecology 11, 2285-2293. Pedersen AA, Loeschcke V (2001) Conservation genetics of peripheral populations of the mygalomorph spider Atypus affinis (Atypidae) in northern Europe. Molecular Ecology 10, 1133-1142. Peichel CL, Nereng KS, Ohgi KA, et al. (2001) The genetic architecture of divergence between threespine stickleback species. Nature 414, 901-905. Peripato AC, de Brito RA, Vaughn TT, et al. (2002) Quantitative trait loci for maternal performance for offspring survival in mice. Genetics 162, 1341-1353. 59 Pfau RS, Van den Bussche RA, McBee K (2001) Population genetics of the hispid cotton rat (Sigmodon hispidus): patterns of genetic diversity at the major histocompatibility complex. Molecular Ecology 10, 1939-1945. Pfeiler E, Markow TA (2001) Ecology and population genetics of Sonoran Desert Drosophila. Molecular Ecology 10, 1787-1791. Pfrender ME, Spitze K, Lehman N (2000) Multi-locus genetic evidence for rapid ecologically based speciation in Daphnia. Molecular Ecology 9, 1717-1735. Pierpaoli M, Riga F, Trocchi V, Randi E (1999) Species distinction and evolutionary relationships of the Italian hare (lepus corsicanus) as described by mitochondrial DNA sequencing. Molecular Ecology 8, 1805-1817. Piertney SB, MacColl ADC, Bacon PJ, Dallas JF (1998) Local genetic structure in red grouse (Lagopus lagopus scoticus): evidence from microsatellite DNA markers. Molecular Ecology 7, 1645-1654. Pinto J, Donnelly MJ, Sousa CA, et al. (2002) Genetic structure of Anopheles gambiae (Diptera : Culicidae) in Sao Tome and Principe (West Africa): implications for malaria control. Molecular Ecology 11, 2183-2187. Pitra C, Lieckfeldt D, Alonso JC (2000) Population subdivision in Europe's great bustard inferred from mitochondrial and nuclear DNA sequence variation. Molecular Ecology 9, 1165-1170. Pope LC, Estoup A, Moritz C (2000) Phylogeography and population structure of an ecotonal marsupial, Bettongia tropica, determined using mtDNA and microsatellites. Molecular Ecology 9, 2041-2053. Pope LC, Sharp A, Moritz C (1996) Population structure of the yellow-footed rock-wallaby Petrogale xanthopus (Gray, 1954) inferred from mtDNA sequences and microsatellite loci. Molecular Ecology 5, 629-640. Procaccini G, Orsini L, Ruggiero MV, Scardi M (2001) Spatial patterns of genetic diversity in Posidonia oceanica, an endemic Mediterranean seagrass. Molecular Ecology 10, 1413-1421. Prosser MR, Gibbs HL, Weatherhead PJ (1999) Microgeographic population genetic structure in the northern water snake, Nerodia sipedon sipedon detected using microsatellite DNA loci. Molecular Ecology 8, 329-333. Prugnolle F, De Meeus T, Durand P, Sire C, Theron A (2002) Sex-specific genetic structure in Schistosoma mansoni: evolutionary and epidemiological implications. Molecular Ecology 11, 1231-1238. Pudovskis MS, Johnson MS, Black R (2001) Genetic divergence of peripherally disjunct populations of the gastropod Batillariella estuarina in the Houtman Abrolhos Islands, Western Australia. Molecular Ecology 10, 2605-2616. Raay TJV, Crease TJ (1995) Mitochondrial DNA diversity in an apomictic Daphnia complex from the Canadian High Arctic. Molecular Ecology 4, 105-114. Ramey II RR (1995) Mitochondrial DNA population structure and evolution of mountain sheep in the south-western United States and Mexico. Molecular Ecology 4, 429-439. Raspe O, Saumitou-Laprade P, Cuguen J, Jacquemart AL (2000) Chloroplast DNA haplotype variation and population differentiation in Sorbus aucuparia L. (Rosaceae : Maloideae). Molecular Ecology 9, 1113-1122. Rassmann R, Tautz D, Trillmich F, Gliddon C (1997) The microevolution of the Galápagos marine iguana Amblyrhynchus cristatus assessed by nuclear and mitochondrial genetic analyses. Molecular Ecology 6, 437-452. Reichow D, Smith MJ (2001) Microsatellites reveal high levels of gene flow among populations of the California squid Loligo opalescens. Molecular Ecology 10, 1101-1109. Rendell S, Ennos RA (2002) Chloroplast DNA diversity in Calluna vulgaris (heather) populations in Europe. Molecular Ecology 11, 69-78. Reusch TBH, Stam WT, Olsen JL (2000) A microsatellite-based estimation of clonal diversity and population subdivision in Zostera marina, a marine flowering plant. Molecular Ecology 9, 127-140. 60 Reusch TBH, Wegner KM, Kalbe M (2001) Rapid genetic divergence in postglacial populations of threespine stickleback (Gasterosteus aculeatus): the role of habitat type, drainage and geographical proximity. Molecular Ecology 10, 2435-2445. Ribeiro MM, Mariette S, Vendramin GG, et al. (2002) Comparison of genetic diversity estimates within and among populations of maritime pine using chloroplast simple-sequence repeat and amplified fragment length polymorphism data. Molecular Ecology 11, 869-877. Richardson BA, Brunsfeld J, Klopfenstein NB (2002) DNA from bird-dispersed seed and wind-disseminated pollen provides insights into postglacial colonization and population genetic structure of whitebark pine (Pinus albicaulis). Molecular Ecology 11, 215-227. Rico C, Turner GF (2002) Extreme microallopatric divergence in a cichlid species from Lake Malawi. Molecular Ecology 11, 1585-1590. Riginos C, Nachman MW (2001) Population subdivision in marine environments: the contributions of biogeography, geographical distance and discontinuous habitat to genetic differentiation in a blennioid fish, Axoclinus nigricaudus. Molecular Ecology 10, 1439-1453. Robin C, Lyman RF, Long AD, Langley CH, Mackay TFC (2002) hairy: A quantitative trait locus for Drosophila sensory bristle number. Genetics 162, 155-164. Robinson NA, Murray ND, Sherwin WB (1993) Vntr Loci Reveal Differentiation between and Structure within Populations of the Eastern Barred Bandicoot Perameles-Gunnii. Molecular Ecology 2, 195-207. Robison BD, Wheeler PA, Sundin K, Sikka P, Thorgaard GH (2001) Composite interval mapping reveals a major locus influencing embryonic development rate in rainbow trout (Oncorhynchus mykiss). Journal of Heredity 92, 16-22. Rocha LA, Bass AL, Robertson DR, Bowen BW (2002) Adult habitat preferences, larval dispersal, and the comparative phylogeography of three Atlantic surgeonfishes (Teleostei : Acanthuridae). Molecular Ecology 11, 243-252. Rodriguez-Lanetty M, Hoegh-Guldberg O (2002) The phylogeography and connectivity of the latitudinally widespread scleractinian coral Plesiastrea versipora in the Western Pacific. Molecular Ecology 11, 1177-1189. Roeder AD, Marshall RK, Mitchelson AJ, et al. (2001) Gene flow on the ice: genetic differentiation among Adelie penguin colonies around Antarctica. Molecular Ecology 10, 1645-1656. Roques S, Duchesne P, Bernatchez L (1999) Potential of microsatellites for individual assignment: the North Atlantic redfish (genus Sebastes) species complex as a case study. Molecular Ecology 8, 1703-1717. Roques S, Sevigny JM, Bernatchez L (2001) Evidence for broadscale introgressive hybridization between two redfish (genus Sebastes) in the North-west Atlantic: a rare marine example. Molecular Ecology 10, 149-165. Rosel PE, France SC, Wang JY, Kocher TD (1999) Genetic structure of harbour porpoise Phocoena phocoena populations in the northwest Atlantic based on mitochondrial and nuclear markers. Molecular Ecology 8, S41-S54. Rosenbaum HC, Brownell RL, Brown MW, et al. (2000) World-wide genetic differentiation of Eubalaena: questioning the number of right whale species. Molecular Ecology 9, 1793-1802. Roslin T (2001) Spatial population structure in a patchily distributed beetle. Molecular Ecology 10, 823-837. Ross TK (1999) Phylogeography and conservation genetics of the Iowa Pleistocene snail. Molecular Ecology 8, 1363-1373. Rossetto M, Slade RW, Baverstock PR, Henry RJ, Lee LS (1999) Microsatellite variation and assessment of genetic structure in tea tree (Melaleuca alternifolia Myrtaceae). Molecular Ecology 8, 633-643. Rossetto M, Weaver PK, Dixon KW (1995) Use of Rapd Analysis in Devising Conservation Strategies for the Rare and Endangered Grevillea-Scapigera (Proteaceae). Molecular Ecology 4, 321-329. Rossiter SJ, Jones G, Ransome RD, Barrattt EM (2000) Genetic variation and population structure in the endangered greater horseshoe bat Rhinolophus ferrumequinum. Molecular Ecology 9, 1131-1135. 61 Rowe G, Beebee TJC, Burke T (1998) Phylogeography of the natterjack toad Bufo calamita in Britain: genetic differentiation of native and translocated populations. Molecular Ecology 7, 751-760. Roy BA, Stanton ML, Eppley SM (1999) Effects of environmental stress on leaf hair density and consequences for selection. Journal of Evolutionary Biology 12, 1089-1103. Ruber L, Meyer A, Sturmbauer C, Verheyen E (2001) Population structure in two sympatric species of the Lake Tanganyika cichlid tribe Eretmodini: evidence for introgression. Molecular Ecology 10, 1207-1225. Russell JR, Weber JC, Booth A, et al. (1999) Genetic variation of Calycophyllum spruceanum in the Peruvian Amazon Basin, revealed by amplified fragment length polymorphism (AFLP) analysis. Molecular Ecology 8, 199-204. Ruzzante DE, Hansen MM, Meldrup D (2001) Distribution of individual inbreeding coefficients, relatedness and influence of stocking on native anadromous brown trout (Salmo trutta) population structure. Molecular Ecology 10, 2107-2128. Ruzzante DE, Taggart CT, Cook D (1998) A nuclear DNA basis for shelf- and bank-scale population structure in northwest Atlantic cod (Gadus morhua): Labrador to Georges Bank. Molecular Ecology 7, 1663-1680. Salvato P, Battisti A, Concato S, et al. (2002) Genetic differentiation in the winter pine processionary moth (Thaumetopoea pityocampa wilkinsoni complex), inferred by AFLP and mitochondrial DNA markers. Molecular Ecology 11, 2435-2444. Sander T, Konig S, Rothe GM, Janssen A, Weisgerber H (2000) Genetic variation of European beech (Fagus sylvatica L.) along an altitudinal transect at mount Vogelsberg in Hesse, Germany. Molecular Ecology 9, 1349-1361. Sarre S (1995) Mitochondrial DNA variation among populations of Oedura reticulata (Gekkonidae) in remnant vegetation: implications for metapopulation structure and population decline. Molecular Ecology 4, 429-439. Saville BJ, Yoell H, Anderson JB (1996) Genetic exchange and recombination in populations of the root-infecting fungus Armillaria gallica. Molecular Ecology 5, 485-497. Schierenbeck KA, Skupski M, Lieberman D, Lieberman M (1997) Population structure and genetic diversity in four tropical tree species in Costa Rica. Molecular Ecology 6, 137-144. Schneider CJ (1996) Distinguishing between primary and secondary intergradation among morphologically differentiated populations of Anolis marmoratus. Molecular Ecology 5, 239-249. Schönswetter P, Tribsch A, Barfuss M, Niklfeld H (2002) Several Pleistocene refugia detected in the high alpine plant Phyteuma globulariifolium Sternb. & Hoppe (Campanulaceae) in the European Alps. Molecular Ecology 11, 2637-2647. Schulte-Hostedde AI, Gibbs HL, Millar JS (2001) Microgeographic genetic structure in the yellow-pine chipmunk (Tamias amoenus). Molecular Ecology 10, 1625-1631. Schultheis AS, Weigt LA, Hendricks AC (2002) Gene flow, dispersal, and nested clade analysis among populations of the stonefly Peltoperla tarteri in the southern Appalachians. Molecular Ecology 11, 317-327. Scotti I, Vendramin GG, Matteotti LS, et al. (2000) Postglacial recolonization routes for Picea abies K. in Italy as suggested by the analysis of sequencecharacterized amplified region (SCAR) markers. Molecular Ecology 9, 699-708. Sebastiani F, Meiswinkel R, Gomulski LM, et al. (2001) Molecular differentiation of the Old World Culicoides imicola species complex (Diptera, Ceratopogonidae), inferred using random amplified polymorphic DNA markers. Molecular Ecology 10, 1773-1786. Seddon JM, Baverstock PR (1999) Variation on islands: major histoincompatibility complex (Mhc) polymorphism in populations of the Australian bush rat. Molecular Ecology 8, 2071-2079. Segelbacher G, Storch I (2002) Capercaillie in the Alps: genetic evidence of metapopulation structure and population decline. Molecular Ecology 11, 1669-1677. 62 Shaffer G, Fellers GM, Magee A, Voss R (2000) The genetics of amphibian declines: population substructure and molecular differentiation in the Yosemite Toad, Bufo canorus (Anura, Bufonidae) based on single-strand conformation polymorphism analysis (SSCP) and mitochondrial DNA sequence data. Molecular Ecology 9, 245-257. Shapcott A (1998) The patterns of genetic diversity in Carpentaria acuminata (Arecaceae), and rainforest history in northern Australia. Molecular Ecology 7, 833-847. Shapcott A (1999) Comparison of the population genetics and densities of five Pinanga palm species at Kuala Belalong, Brunei. Molecular Ecology 8, 16411654. Shaw KL, Parsons YM (2002) Divergence of mate recognition behavior and its consequences for genetic architectures of speciation. American Naturalist 159, S61-S75. Shaw PW, Pierce GJ, Boyle PR (1999) Subtle population structuring within a highly vagile marine invertebrate, the veined squid Loligo forbesi, demonstrated with microsatellite DNA markers. Molecular Ecology 8, 407-417. Shook DR, Johnson TE (1999) Quantitative trait loci affecting survival and fertility- related traits in Caenorhabditis elegans show genotype- environment interactions, pleiotropy and epistasis. Genetics 153, 1233-1243. Silva C, Eguiarte LE, Souza V (1999) Reticulated and epidemic population genetic structure of Rhizobium etli biovar phaseoli in a traditionally managed locality in Mexico. Molecular Ecology 8, 277-287. Simon JC, Baumann S, Sunnucks P, et al. (1999) Reproductive mode and population genetic structure of the cereal aphid Sitobion avenae studied using phenotypic and microsatellite markers. Molecular Ecology 8, 531-545. Simonsen BT, Siegismund HR, Arctander P (1998) Population structure of African buffalo inferred from mtDNA sequences and microsatellite loci: high variation and low differentiation. Molecular Ecology 7, 225-237. Sinclair WT, Morman JD, Ennos RA (1999) The postglacial history of Scots pine (Pinus sylvestris L.) in western Europe: evidence from mitochondrial DNA variation. Molecular Ecology 8, 83-88. Skotnicki ML, Ninham JA, Selkirk PM (1999) Genetic diversity and dispersal of the moss Sarconeurum glaciale on Ross Island, East Antarctica. Molecular Ecology 8, 753-762. Small MP, Beacham TD, Withler RE, Nelson RJ (1998) Discriminating coho salmon (Oncorhynchus kisutch) populations within the Fraser River, British Columbia, using microsatellite markers. Molecular Ecology 7, 141-155. Small MP, Gosling EM (2000) Species relationships and population structure of Littorina saxatilis Olivi and L. tenebrosa Montagu in Ireland using single-strand conformational polymorphisms (SSCPs) of cytochrome b fragments. Molecular Ecology 9, 39-52. Soranzo N, Alia R, Provan J, Powell W (2000) Patterns of variation at a mitochondrial sequence-tagged-site locus provides new insights into the postglacial history of European Pinus sylvestris populations. Molecular Ecology 9, 1205-1211. Stacy JE, Jordan PE, Steen H, et al. (1997) Lack of concordance between mtDNA gene flow and population density fluctuations in the bank vole. Molecular Ecology 6, 751-759. Stangel PW, Lennartz MR, Smith MH (1992) Genetic variation and population structure of red-cockaded woodpeckers. Conservation Biology 6, 283-292. Stehlik I, Schneller JJ, Bachmann K (2001) Resistance or emigration: response of the high-alpine plant Eritrichium nanum (L.) Gaudin to the ice age within the Central Alps. Molecular Ecology 10, 357-370. Steinger T, Haldimann P, Leiss KA, Müller-Schärer H (2002) Does natural selection promote population divergence? A comparative analysis of population structure using amplified fragment length polymorphism markers and quantitative traits. Molecular Ecology 11, 2583-2590. 63 Stenlid J, Vasiliauskas R (1998) Genetic diversity within and among vegetative compatibility groups of Stereum sanguinolentum. Molecular Ecology 7, 12651274. Stenstrom A, Jonsson BO, Jonsdottir IS, Fagerstrom T, Augner M (2001) Genetic variation and clonal diversity in four clonal sedges (Carex) along the Arctic coast of Eurasia. Molecular Ecology 10, 497-513. Stepien AA, Faber JE (1998) Population genetic structure, phylogeography and spawning philopatry in walleye (Stizostedion vitreum) from mitochondrial control region sequences. Molecular Ecology 7, 1757-1769. Stepien CA (1999) Phylogeographical structure of the Dover sole Microstomus pacificus: the larval retention hypothesis and genetic divergence along the deep continental slope of the northeastern Pacific Ocean. Molecular Ecology 8, 923-939. Stevens MT, Turner MG, Tuskan GA, et al. (1999) Genetic variation in postfire aspen seedlings in Yellowstone National Park. Molecular Ecology 8, 1769-1780. Strain SR, Whittam TS, Bottomley PS (1995) Analysis of genetic structure in soil populations of Rhizobium leguminosarum recovered from the USA and the UK. Molecular Ecology 4, 105-114. Sucena E, Stern DL (2000) Divergence of larval morphology between Drosophila sechellia and its sibling species caused by cis-regulatory evolution of ovo/shaven-baby. Proceedings of the National Academy of Sciences of the United States of America 97, 4530-4534. Sun M (1999) Cleistogamy in Scutellaria indica (Labiatae): effective mating system and population genetic structure. Molecular Ecology 8, 1285-1295. Takahashi A, Tsaur SC, Coyne JA, Wu CI (2001) The nucleotide changes governing cuticular hydrocarbon variation and their evolution in Drosophila melanogaster. Proceedings of the National Academy of Sciences of the United States of America 98, 3920-3925. Tarr CL, Conant S, Fleischer RC (1998) Founder events and variation at microsatellite loci in an insular passerine bird, the Laysan finch (Telespiza cantans). Molecular Ecology 7, 719-731. Tarr CL, Fleischer RC (1999) Population boundaries and genetic diversity in the endangered Mariana crow (Corvus kubaryi). Molecular Ecology 8, 941-949. Taylor AC, Sherman WB, Wayne RK (1994) Genetic variation of microsatellite loci in a bottlenecked species: the northern hairy-nosed wombat Lasiorhinus kreftii. Molecular Ecology 3, 277-290. Taylor EB, Harvey S, Pollad S, Volpe J (1997) Postglacial genetic differentiation of reproductive ecotypes of kokanee Oncorhynchus nerka in Okanagan Lake, British Columbia. Molecular Ecology 6, 503-517. Taylor EB, Pollard S, Louie D (1999) Mitochondrial DNA variation in bull trout (Salvelinus confluentus) from northwestern North America: implications for zoogeography and conservation. Molecular Ecology 8, 1155-1170. Tek Tay W, Cook JM, Rowe DJ, Crozier RH (1997) Migration between nests in the Australian arid-zone ant Rhytidoponera sp. 12 revealed by DGGE analysis of mitochondrial DNA. Molecular Ecology 6, 403-411. Tessier N, Bernatchez L (1999) Stability of population structure and genetic diversity across generations assessed by microsatellites among sympatric populations of landlocked Atlantic salmon (Salmo salar L.). Molecular Ecology 8, 169-179. Tessier N, Bernatchez L, Wright JM (1997) Population structure and impact of supportive breeding inferred from mitochondrial and microsatellite DNA analyses in land-locked Atlantic salmon Salmo salar L. Molecular Ecology 6, 735-750. Thurston HD (1961) The relative survival ability of races of Phytophthora infestans in mixtures. Phytopathology 51, 748-755. Toline CA, Baker AJ (1995) Mitochondrial DNA variation and population genetic structure of the northern redbelly dace (Phoxinus eos). Molecular Ecology 4, 745-753. Tollefsrud MM, Bachmann K, Jakobsen KS, Brochmann C (1998) Glacial survival does not matter - II: RAPD phylogeography of Nordic Saxifraga cespitosa. Molecular Ecology 7, 1217-1232. 64 Travis SE, Maschinski J, Keim P (1996) An analysis of genetic variation in Astragalus cremnophylax var cremnophylax a critically endangered plant, using AFLP markers. Molecular Ecology 5, 735-745. Tremblay NO, Schoen DJ (1999) Molecular phylogeography of Dryas integrifolia: glacial refugia and postglacial recolonization. Molecular Ecology 8, 11871198. Triantafyllidis A, Krieg F, Cottin C, et al. (2002) Genetic structure and phylogeography of European catfish (Silurus glanis) populations. Molecular Ecology 11, 1039-1055. True JR, Liu JJ, Stam LF, Zeng ZB, Laurie CC (1997) Quantitative genetic analysis of divergence in male secondary sexual traits between Drosophila simulans and Drosophila mauritiana. Evolution 51, 816-832. Tsagkarakou S, Navajas M, Papaioannou-Souliotis P, Pasteur N (1998) Gene flow among Tetranychus urticae (Acari: Tetranychidae) populations in Greece. Molecular Ecology 7, 71-79. Turpeinen T, Tenhola T, Manninen O, Nevo E, Nissila E (2001) Microsatellite diversity associated with ecological factors in Hordeum spontaneum populations in Israel. Molecular Ecology 10, 1577-1591. Uphyrkina O, Johnson WE, Quigley H, et al. (2001) Phylogenetics, genome diversity and origin of modern leopard, Panthera pardus. Molecular Ecology 10, 2617-2633. Van de Zande L, Van Apeldoorn RC, Blijdenstein AF, et al. (2000) Microsatellite analysis of population structure and genetic differentiation within and between populations of the root vole, Microtus oeconomus in the Netherlands. Molecular Ecology 9, 1651-1656. Van der Strate HJ, Van de Zande L, Stam WT, Olsen JL (2002) The contribution of haploids, diploids and clones to fine-scale population structure in the seaweed Cladophoropsis membranacea (Chlorophyta). Molecular Ecology 11, 329-345. van Hooft WF, Groen AF, Prins HHT (2000) Microsatellite analysis of genetic diversity in African buffalo (Syncerus caffer) populations throughout Africa. Molecular Ecology 9, 2017-2025. van Tienderen PH, van der Toorn J (1991) Genetic Differentiation between Populations of Plantago- Lanceolata .2. Phenotypic Selection in a Transplant Experiment in 3 Contrasting Habitats. Journal of Ecology 79, 43-59. Van Vuuren BJ, Robinson TJ (1997) Genetic population structure in the yellow mongoose, Cynictis penicillata. Molecular Ecology 6, 1147-1153. Vandewoestijne S, Neve G, Baguette M (1999) Spatial and temporal population genetic structure of the butterfly Aglais urticae L-(Lepidoptera, Nymphalidae). Molecular Ecology 8, 1539-1543. Vanlerberghe-Masuti F, Chavigny P (1998) Host-based genetic differentiation in the aphid Aphis gossypii Glover, evidence from RAPD fingerprints. Molecular Ecology 7, 905-914. Vaughn TT, Pletscher LS, Peripato A, et al. (1999) Mapping quantitative trait loci for murine growth: a closer look at genetic architecture. Genetical Research 74, 313-322. Vendramin GG, Degen B, Petit RJ, et al. (1999) High level of variation at Abies alba chloroplast microsatellite loci in Europe. Molecular Ecology 8, 1117-1126. Vernesi C, Pecchioli E, Caramelli D, et al. (2002) The genetic structure of natural and reintroduced roe deer (Capreolus capreolus) populations in the Alps and central Italy, with reference to the mitochondrial DNA phylogeography of Europe. Molecular Ecology 11, 1285-1297. Vieira C, Pasyukova EG, Zeng ZB, et al. (2000) Genotype-environment interaction for quantitative trait loci affecting life span in Drosophila melanogaster. Genetics 154, 213-227. Vilá C, Amorim IR, Leonard JA, et al. (1999) Mitochondrial DNA phylogeography and population history of the grey wolf Canis lupus. Molecular Ecology 8, 2089-2103. 65 von Segesser F, Menard N, Gaci B, Martin RD (1999) Genetic differentiation within and between isolated Algerian subpopulations of Barbary macaques (Macaca sylvanus): evidence from microsatellites. Molecular Ecology 8, 433-442. Vucetich LM, Vucetich JA, Joshi CP, Waite TA, Peterson RO (2001) Genetic (RAPD) diversity in Peromyscus maniculatus populations in a naturally fragmented landscape. Molecular Ecology 10, 35-40. Waits L, Taberlet P, Swenson JE, Sandegren F, Franzen R (2000) Nuclear DNA microsatellite analysis of genetic diversity and gene flow in the Scandinavian brown bear (Ursus arctos). Molecular Ecology 9, 421-431. Walker CW, Vila C, Landa A, Linden M, Ellegren H (2001) Genetic variation and population structure in Scandinavian wolverine (Gulo gulo) populations. Molecular Ecology 10, 53-+. Walter R, Epperson BK (2001) Geographic pattern of genetic variation in Pinus resinosa: area of greatest diversity is not the origin of postglacial populations. Molecular Ecology 10, 103-111. Walton C, Handley JM, Collins FH, et al. (2001) Genetic population structure and introgression in Anopheles dirus mosquitoes in South-east Asia. Molecular Ecology 10, 569-580. Wang H-Y, Tsai M-P, Yu M-J, Lee S-C (1999) Influence of glaciation on divergence patterns of the endemic minnow, Zacco pachycephalus, in Taiwan. Molecular Ecology 8, 1879-1888. Wang JP, Hsu KC, Chiang TY (2000) Mitochondrial DNA phylogeography of Acrossocheilus paradoxus (Cyprinidae) in Taiwan. Molecular Ecology 9, 14831494. Warner RR, Schultz ET (1992) Sexual Selection and Male Characteristics in the Bluehead Wrasse, Thalassoma-Bifasciatum - Mating Site Acquisition, Mating Site Defense, and Female Choice. Evolution 46, 1421-1442. Watanabe Y, Chiba S (2001) High within-population mitochondrial DNA variation due to microvicariance and population mixing in the land snail Euhadra quaesita (Pulmonata : Bradybaenidae). Molecular Ecology 10, 2635-2645. Waters JM, Dijkstra LH, Wallis GP (2000) Biogeography of a southern hemisphere freshwater fish: how important is marine dispersal? Molecular Ecology 9, 1815-1821. Wayne ML, Hackett JB, Dilda CL, et al. (2001) Quantitative trait locus mapping of fitness-related traits in Drosophila melanogaster. Genetical Research 77, 107-116. Weiss S, Antunes A, Schlotterer C, Alexandrino P (2000) Mitochondrial haplotype diversity among Portuguese brown trout Salmo trutta L. populations: relevance to the post-Pleistocene recolonization of northern Europe. Molecular Ecology 9, 691-698. Welch ME, Gerber GP, Davis SK (in press) The genetic structure of the Turks and Caicos rock iguana, Cyclura carinata carinata, and its implications for species conservation. Welz HG, Leonard KJ (1993) Phenotypic Variation and Parasitic Fitness of Races of Cochliobolus-Carbonum on Corn in North-Carolina. Phytopathology 83, 593-601. Wenburg JK, Bentzen P, Foote CJ (1998) Microsatellite analysis of genetic population structure in an endangered salmonid: the coastal cutthroat trout (Oncorhynchus clarki clarki). Molecular Ecology 7, 733-749. Wenink PW, Groen AF, Roelke-Parker ME, Prins HHT (1998) African buffalo maintain high genetic diversity in the major histocompatibility complex in spite of historically known population bottlenecks. Molecular Ecology 7, 1315-1322. Weston RF, Qureshi I, Werren JH (1999) Genetics of wing size differences between two Nasonia species. Journal of Evolutionary Biology 12, 586-595. White G, Powell W (1997) Isolation and characterization of microsatellite loci in Swietenia humilis (Meliaceae): an endangered tropical hardwood species. Molecular Ecology 6, 851-860. 66 White GM, Boshier DH, Powell W (1999) Genetic variation within a fragmented population of Swietenia humilis Zucc. Molecular Ecology 8, 1899-1909. Whitehouse AM, Harley EH (2001) Post-bottleneck genetic diversity of elephant populations in South Africa, revealed using microsatellite analysis. Molecular Ecology 10, 2139-2149. Widmer A, Schmid-Hempel P (1999) The population genetic structure of a large temperate pollinator species, Bombus pascuorum (Scopoli) (Hymenoptera : Apidae). Molecular Ecology 8, 387-398. Williams MA, Blouin AG, Noor MAF (2001) Courtship songs of Drosophila pseudoobscura and D-persimilis. II. Genetics of species differences. Heredity 86, 68-77. Williams ST, Benzie JAM (1997) Indo-West Pacific patterns of genetic differentiation in the high-dispersal starfish Linckia laevigata. Molecular Ecology 6, 559573. Wilson AB, Boates JS, Snyder M (1997) Genetic isolation of the gammaridean amphipod, Corophium volutator, in the Bay of Fundy, Canada. Molecular Ecology 6, 917-923. Wolf JB, Vaughn TT, Pletscher LS, Cheverud JM (2002) Contribution of maternal effect QTL to genetic architecture of early growth in mice. Heredity 89, 300310. Wolff K, ElAkkad S, Abbott RJ (1997) Population substructure in Alkanna orientalis (Boraginaceae) in the Sinai desert, in relation to its pollinator behaviour. Molecular Ecology 6, 365-372. Workman MS, Leamy LJ, Routman EJ, Cheverud JM (2002) Analysis of quantitative trait locus effects on the size and shape of mandibular molars in mice. Genetics 160, 1573-1586. Wynen LP, Goldsworthy SD, Guinet C, et al. (2000) Postsealing genetic variation and population structure of two species of fur seal (Arctocephalus gazella and A-tropicalis). Molecular Ecology 9, 299-314. Xu J, Kerrigan RW, Sonnenberg AS, et al. (1998) Mitochondrial DNA variation in natural populations of the mushroom Agaricus bisporus. Molecular Ecology 7, 19-13. Yan G, Romero-Severson J, Walton M, Chadee DD, Severson DW (1999) Population genetics of the yellow fever mosquito in Trinidad: comparisons of amplified fragment length polymorphism (AFLP) and restriction fragment length polymorphism (RFLP) markers. Molecular Ecology 8, 951-963. Young DL, Allard MW (1997) Conservation genetics of the plain pigeon Columbia inornata in Puerto Rico and the Dominican Republic. Molecular Ecology 8, 877-879. Zawko G, Krauss SL, Dixon KW, Sivasithamparam K (2001) Conservation genetics of the rare and endangered Leucopogon obtectus (Ericaceae). Molecular Ecology 10, 2389-2396. Zeng ZB, Liu JJ, Stam LF, et al. (2000) Genetic architecture of a morphological shape difference between two drosophila species. Genetics 154, 299-310. Zoller S, Lutzoni F, Schneider C (1999) Genetic variation within and among populations of the threatened lichen Lobaria pulmonaria in Switzerland and implications for its conservation. Molecular Ecology 8, 2049-2069.








