README.
advertisement

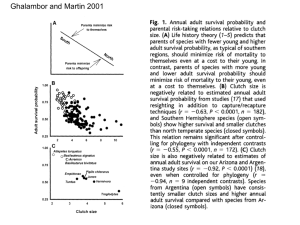
Data description growth.txt Size data for adult Crepidula fornicata from 25 stacks that were reared in the lab during 251 days (January 12 to September 20, 2011). Data are given only for the individuals that were alive at day 251 (n=175 individuals out of 223; data were missing for 3 individuals). Each line corresponds to one individual. Stacks are labeled from 9 to 33 (stacks 1 to 8 were not used for growth analysis, see Table 1 in manuscript). Columns "size" and "size2" give straight shell length estimated at day 1 (January 12, 2011) and day 251 (September 20, 2011). "Growth" is the difference between this numbers. It can be negative because there is some uncertainty associated with the measurements taken when the individuals were alive (it is more difficult to measure size when the stacks are not dismantled). "Position" is the position of each individual within a stack, starting from the base (i.e. individual in position 1 is usually the basal female of a stack). The sign">x" sign means that the corresponding individual was attached on the side of individual x (see Fig. 3 in Dupont et al. 2006). genotypes_adults.txt This file contains the genotypes at 8 microsatellite loci for the adults from stacks 1 to 8 (n=69, the genotypes from individuals S3A5 and S8A15 are missing). Individual labels are of the form SxAy with x the stack identity and y individual identity within this stack. genotypes_larvae.txt This file contains the genotypes at 8 microsatellite loci for a sample of the larvae produced by stacks 1 to 8 (n=3240 larvae with at least 5 loci successfully typed). Each larva comes from a given larval release (column "release", see main text for definition). fecundity.txt Estimates of brood size (total number of larvae) for 44 broods produced by 24 females from stacks 1 to 8 (see Table 1 in manuscript). Each line corresponds to one brood (that is, produced by a single female, as defined after parentage assignment, see main text for a definition of this term). In general, several broods were analyzed per female. Columns "Nlarvae" and "sd" give estimates of the total number of larvae in a brood and standard deviation (estimated from 6 samples per brood, see main text). Standard deviations were not calculated when a brood was sampled simultaneously with another one (i.e. one observation of a larval release contained two broods, see main text) or when it was distributed across two larval releases. sire_contributions.txt This file gives the number of larvae in a sample that were sired by a given father. Each line corresponds to a fraction of a brood produced by a unique mother and a unique father. The first column is the label of the stack (stacks 1-8, see Table 1 in article). Column "brood" is the label of the brood analyzed. See the definition of brood in the article. Because broods can be defined only after genetic parentage assignment, brood labelling is as follows: brood x corresponds to larval release x (see main text), broods xa and xb correspond to two broods (from distinct mothers) that were identified from a unique larval release x (i.e. synchronous production of larvae by two females), brood x_y correspond to a unique brood that was sampled from two separate larval releases (larvae released by the same mother with a small time interval, see main text). One brood has a unique mother but can have several fathers, given in columns "father" and "mother". The number of larvae assigned to each particular parental pair (one mother and one father) is given in column "n". Column "ntot" is the total number of larvae genetically identified as belonging to the relevant brood. Hence n/ntot gives the proportion of a brood sired by a given father (column "prop"). Column "dist" is the distance between the father and the mother (see main text). Column "sex" gives the sex of the father (which can be female in case of sperm storage). reproductive_success.txt Reproductive success data for each individual from the 8 stacks listed in table 1: number of male and female mating partners (columns "male_function" and "female_function"), total mating success (=total number of mating partners, in column "W1tot"), reproductive success through the male and female functions ("Wfather" and "Wmother") and total reproductive success ("Wtot"). The sex and size of individuals are also provided. References Dupont, L., J. Richard, Y.-M. Paulet, G. Thouzeau, and F. Viard. 2006. Gregariousness and protandry promote reproductive insurance in the invasive gastropod Crepidula fornicata: evidence from assignment of larval paternity. Mol. Ecol. 15:3009-3021.