Mol bioinf / SVarvio / EXAM / 14. Oct 2010 / C128 Computer class
advertisement

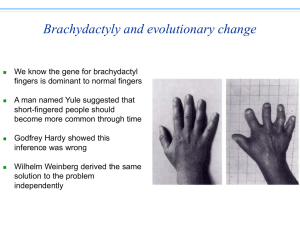
Mol bioinf / SVarvio / EXAM / 14. Oct 2010 / C128 Computer class Send answers to sirkka-liisa.varvio@helsinki.fi, use .doc, .txt or .pdf. Include your student number. Before leaving the classroom: check that your e-mail has been received. 1. (25 points) University of Helsinki page News&Views, a couple of days ago: http://www.helsinki.fi/news/archive/10-2010/12-13-19-14.html. “The Michael J. Fox Foundation for Parkinson’s Research has awarded a grant worth in excess of half a million dollars to a research group of Academy professor Mart Saarma of the Institute of Biotechnology, Viikki, for the development of a new treatment for Parkinson’s disease. The treatment under development is based on the CDNF growth factor discovered by Saarma's group.” a) When and where (in what journal(s)) has Saarma´s group published their research about this gene. Give reference(s)? b) Has some other research group also published about this gene? c) What information you can find about this gene from sequence databases? d) If you want to compare human CDNF sequence with animal sequences, what are the animal species from which full length (as compared with the human gene sequence information) exist? Give also names of some animals from which less than “full length” (as compared with the human sequence) sequence information is available. e) Describe the necessary steps and procedures you need to take in order to construct a neighbor joining phylogeny from the available information of this gene. 2. (5 points) You know that in the Hardy-Weinberg “equilibrium” the allele frequencies and genotypes frequencies have a certain relationship (Two alleles with frequencies p and q, three genotypes with frequencies p2 and 2pq and q2 (p + q = 1 and p2 + 2pq + q2 = 1). What about a case in which there are three alleles, p + q + r = 1. What are the Hardy-Weinberg genotype frequencies? Human ABO blood group system has three major alleles (in addition more than 70 molecularly distinct alleles): IA, IB and IO. Allele IO is recessive as regards alleles IA and IB (IO gene has a single nucleotide deletion which results in inactive protein). Genotypes IAIA and IAIO are phenotypically A (the blood group A), IBIB and IBIO are phenotypically B (the blood group B), IAIB is phenotypically AB and IO IO is O (the blood group O). Allele frequencies in one human population are: 0.3 for IA, 0.6 for IB and 0.1 for IO. Assuming Hardy-Weinberg equilibrium, what are the blood group phenotype frequencies in the population? 3. (2 points) Explain the connection of bootstrapping and phylogenetic tree topology. Why is bootstrapping performed? 4. (2 points) Four OTU sequences are the material for maximum parsimony and for maximum likelihood phylogeny inference analysis. Focus is in one particular nucleotide site (as in the examples in lecture slides). Why is it that many more possibilities must be considered for maximum likelihood procedure than for parsimony procedure? 5. (2 points) The parameter assumptions in Jukes-Cantor model are unrealistic. What do you think Thomas Jukes and Charles Cantor had in mind when they derived the model, why did they make such a simple model? 6. (2 points) Why is mitochondrial DNA a very good marker molecule for population differences and for evolutionary tracing of human population histories? 7. (2 points) Go to NCBI site and find a page in which the relationships between different databases are presented. How many links are there from SNP-database to nucleotide-database?