Theoretical base for comparison of the mRNA levels between
advertisement

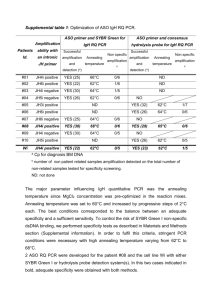
1 1 Supplementary Materials 2 3 Methods 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 Theoretical base for comparison of the mRNA levels between different enzymes and receptors We used the method of comparison in order to compare mRNA levels of different enzymes/receptors, which were obtained by using different primers, as described in (Kimoto et al., 2010). The following consideration is necessary. As an example, we compare AR (Ar) in CC at 24 m and that of P450arom (Cyp19a1) in the hippocampus (Hi) at 3 m. Since we use P450arom in 3m Hi as a standard, we obtain NAr CC / NCyp19a1 Hi (1) where ‘N’ is the number of mRNA molecules and we assumed that we obtain the ng i molecules of cDNA from Ng i molecules of mRNA via RT with an equal efficacy in the same sample. Therefore, in the process of RT, we have the following relationship nAr CC / NAr CC = nGapdh CC / NGapdh CC (2) nCyp19a1 Hi / NCyp19a1 Hi = nGapdh Hi / NGapdh Hi (3) where Gapdh is a house-keeping standard gene which is assumed to express at equal level in these organs (NGapdh CC = NGapdh Hi). In Fig.1, we obtained nGapdh CC = nGapdh Hy = nGapdh CL = nGapdh Hi, indicating that Gapdh is a good internal standard gene. Finally we obtain NAr CC / NCyp19a1 Hi = (nAr CC / nGapdh CC) / (nCyp19a1 Hi / nGapdh Hi) (4) Then we need to obtain ‘nAr CC / nGapdh CC’ from the EtBr band intensity ‘I’. The number of cDNA molecules, ‘n’, is exponentially amplified by PCR, and its PCR product contains ‘n × L ×(1+e)c ’ molecules, where ‘c’ is a PCR cycle number, ‘L’ is a product length, and ‘e’ is an amplification efficacy obtained from the PCR amplification curves in the exponential amplification phase (linear phase with semi-log plot). The band intensity of the PCR product ‘I’ is proportional to the molecule numbers. Therefore, we can obtain ‘n’ from the next equation: I = A× n × L × (1+e)c (5) where ‘A’ is a proportionality coefficient between ‘I’ and ‘n × L ×(1+e)c’. From amplification curves, we choose the exponential amplification phase of PCR cycles which satisfy the equation (5). The PCR cycle in the exponential phase was 24-30, the product length (LAr CC) was 536 for 17β-HSD type 3 in the CC (Table S1) and we chose cAr CC = 26 and determined eAr CC = 1.0850, by fitting the data of ‘c’ and ‘I’ to the equation (5) (I = 0.87 × 2.0850c, R2 = 0.9037). For the Gapdh bands, the same 2 37 38 39 40 41 42 43 calculation was performed, and then we obtained cGapdh CC = 18 and eGapdh CC = 0.9310 (I = 17.53 × 1.9310c, R2 = 0.9295). From these values we can calculate ‘n’ from the band intensity ‘I’. nArCC / nGapdh CC = (IArCC / IGapdh CC) × (LGapdh CC / LArCC) × (1+0.9310)18 / (1+1.0850)26 (6) By substitution eq. (6) into eq. (4), we can compare the mRNA levels for different enzymes and receptors.