Additional file 1
advertisement

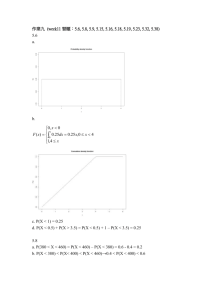
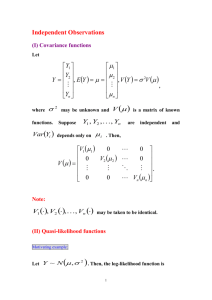
Additional file 1: Joint test of main and interaction effects Let Y = 0,1 indicate control and case status, respectively; let X be the vector of covariates including treatment assignment in the WHI trials, potential confounders (log transformed Gail 5-year breast cancer risk score, previous hormone use for each of estrogen and estrogen plus progestin, log transformed body mass index), variables used for matching controls to cases (baseline age, self-reported ethnicity, participation in each trial component, years since randomization, and baseline hysterectomy status), and first ten principal components for the SNP genotype data. For a particular SNP, let G = 0,1,2 indicate the number of minor alleles. The following five logistic regression models are applied to case-control or case-only samples: (0) Applying a logistic regression model of Y on G and X to the case-control sample: log Pr(Y 1 | G, X ) 0 1G 2T X 1 Pr(Y 1 | G, X ) and let ̂ 1 be the estimator of 1 . (1 to 4) Let Zk = 0,1; k = 1,2,3,4 as the indicator that a subject is assigned to the placebo or active arm of the E-alone trial, E+P trial, DMQ trial, and CaD trial respectively. Applying a logistic regression model to all the cases in the corresponding trial component: log where log Pr( Z k 1 | G) q log k k k G, 1 Pr( Z k 1 | G) 1 qk qk is the offset term, with qk the fraction of women assigned to the 1 qk active arm in the kth trial component. Denote corresponding parameter estimates to be ˆ k , ˆk . A chi-square test with 5 degrees of freedom is used to test the hypothesis that 1 , 1 , 2 , 3 , 4 T 0 0 0 0 0T . Specifically, we reject the null hypothesis if T t (ˆ) var(ˆ) 1ˆ 52,0.95 , where ˆ ˆ1 , ˆ1 , ˆ2 , ˆ3 , ˆ4 , 52,0.95 is the lower 95th percentile of chi-square distribution with 5 degrees of freedom, and t(.) denotes vector transpose. As shown in [23], ̂ 1 is asymptotically independent of [ˆ1 , ˆ2 , ˆ3 , ˆ4 ], ; thus, the offdiagonal elements in the first row and first column of var(ˆ) will all be zero and ˆ1 ˆ 2 t (ˆ)V (ˆ) 1ˆ ˆ 1 / var(ˆ 1 ) [ˆ1 , ˆ 2 , ˆ3 , ˆ 4 ] var[ ˆ1 , ˆ 2 , ˆ3 , ˆ 4 ] 1 2 . ˆ3 ˆ 4 Note that var(ˆ ) can be obtained from the logistic model fitting in (0), and we estimate var[ˆ1 , ˆ2 , ˆ3 , ˆ4 ] using a sandwich type estimator. For subject i in the case-control sample, let U1i, U2i, U3i, U4i be the score function for Z ki E ( Z k | Gi ) models 1 to 4, respectively; U ki , k 1,2,3,4 if subject i is a case Gi ( Z ki E ( Z k | Gi )) in trial component k, otherwise Uki = (0,0)T. Let I1, I2, I3, I4 be the information matrix for models 1 to 4, respectively, with dimensions of 4x4 each, and let N be the total sample size of the case-control sample: I k i 1 I ki N where Gi E ( Z k | Gi )(1 E ( Z k | Gi )) E ( Z k | Gi )(1 E ( Z k | Gi )) I ki 2 Gi E ( Z k | Gi )(1 E ( Z k | Gi )) Gi E ( Z k | Gi )(1 E ( Z k | Gi )) if subject i is a case in trial component k, otherwise: 0 0 I ki . 0 0 Then the sandwich estimator for variance of [ˆ1 , ˆ1 , ˆ2 , ˆ2 , ˆ3 , ˆ3 , ˆ4 , ˆ4 ]T can be computed as: I1 0 0 0 0 0 I2 0 0 I3 0 0 0 0 0 I4 1 U 1i N U 2i U 1i i 1 U 3i U 4i U 2i U 3i I1 0 U 4i 0 0 0 0 I2 0 0 I3 0 0 0 0 0 I4 1 from which we can obtain estimate of var[ˆ1 , ˆ2 , ˆ3 , ˆ4 ] . Furthermore, note that since subjects in the E-alone trial and the E+P trial are nonoverlapping, we have ˆ1 ˆ2 , and the covariance term between ˆ1 and ˆ 2 in the estimate of var[ˆ1 , ˆ2 , ˆ3 , ˆ4 ] is set equal to zero.