Swiss PDB Viewer exercises & answers
advertisement

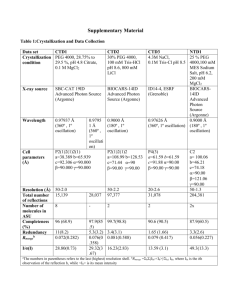
Swiss PDB Viewer exercises & answers General instructions Save your answers into a separate Word file for your own reference. To import images from Swiss PDB Viewer to Word you can take screenshots and paste them into Word. Activate the desired window in Swiss PDB Viewer by clicking it and press Alt + Print Scrn to take a screen shot of that window. In Word, place the cursor and press Ctrl + v to paste the image. 1. Checking PDB file against its Uniprot entry Cutting parts of the protein is often required for obtaining protein crystals needed in tertiary structure Cutting parts of the protein is often required for obtaining protein crystals needed in tertiary structure determination. Due to this, the sequence numbering in PDB files may be different from the corresponding Uniprot sequences. Find out what is the sequence difference between chicken egg-white lysozyme from Swissprot (LYSC_CHICK) and one of its corresponding sequences in PDB file: 1hew. Instructions: 1. Obtain the sequences for LYSC_CHICK and 1hew in fasta format. Go to the PDB website (http://www.rcsb.org/pdb/). Fetch the entry for 1hew, and Download sequences in the entry in FASTA format. 2. Obtain the sequence of LYSC_CHICK from SRS. 3. Copy and paste the two sequences into SRS global alignment tool called StretcherP. Which part of LYSC_CHICK sequence is missing from the PDB entry? How many residues are missing from 1hew? Answer: 18 N-terminal residues are missing: MRSLLILVLCFLPLAALG 2.Molecular surface and active site analysis For PDB file 1hew (lysozyme in complex with inhibitor) create a view that shows the molecular surface of the protein and the bound enzyme inhibitor in wireframe. Colour the conserved residues identified in yesterday’s exercise 4.2 for the LYSC_CHICK protein in green and the rest of the surface grey. The image should look similar to figure 1. Figure 1 Example showing molecular surface for Myoglobin (1mbn), prosthetic heme group is shown in wireframe. Instructions: 1. First change the setting for molecular surface calculations, so as to allow you to exclude selected residues from the surface. Preferences > Surface > Ignore Selected Residues 2. Select enzyme inhibitor (NAG) from the control panel (you can select several residues by holding the Ctrl-button while selecting the residues with left mouse clicks, or select a continuous stretch of residues by holding down the left mouse button on the first residue and dragging down to the last one you wish to select). 3. Show the selected inhibitor residues in wireframe. 4. Next, compute the surface of the protein. You need the NAG residues selected soas to exclude them from the surface rendering. Tools > Compute Molecular Surface 5. You should also turn on OpenGL rendering see a solid surface. Display > Use OpenGL rendering 6. Colour the surface grey, use the control panel for this. 7. Select conserved residues (see list below) and colour them green You have to change LYSC_CHICK sequence numbering to correspond to 1hew’s numbering!!! Conserved residues in LYSC_CHICK excluding cysteines as identified in yesterday’s exercise 4.2.: W S T G Q I D I W LYSC_CHICK 46 54 58 72 75 76 105 116 126 Tips: Discard molecular surface: File > Discard > Discard Surface Questions: 2.1 2.2 Where are the conserved residues located? Where is the inhibitor molecule bound? W S T G Q I D I W LYSC_CHICK 46 1hew 28 54 36 58 40 72 54 75 57 76 58 105 87 116 98 126 108 The conserved residues are located mainly in a groove on the protein’s surface, where the inhibitor is also bound. This is the active site of the enzyme. 3. Ramachandran plot Ramachandran plots can be used to check that the given/assumed 3D structure of a model is of reasonable quality. For a given protein, the torsion angles (or conformational angles): phi and psi of the polypeptide backbone may be plotted. For example, in the plot below, each dot is the phi/psi angle of one residue. In Swiss PDB Viewer, the Ramachandran plots can be used to modify torsion angle by dragging points on the plot – try it and see the effect on the structure! Beta sheet region Left handed alpha helix region Right handed alpha helix region Questions. 3.1 For protein 1hxw (HIV protease dimer), make a Ramachandran plot and identify the residues which are outside the expected (yellow) regions of the plot. They are almost exclusively one kind of residue – which? Why do you think this residue type is more likely to have greater freedom in backbone torsion angles than other residues? Answer: The residues which are mostly outside the expected regions are glycines – this is because they have no side chain and are therefore the smallest residues, with the least constraints on backbone conformation or position. 4. Analysis of a Protein-DNA complex Obtain the full PDB entry for protein: 1flo, and load the protein into Swiss PDB Viewer. Try and obtain a view like the one below, but with all four hetero atoms. Note that your image should include the following: Only DNA and hetero atoms are shown the backbone is displayed in ribbon form, coloured by secondary structure the heterogeneous atoms are highlighted and solid, the heterogeneous atoms are labelled Hint: Select GroupKind:HETATM will identify the heterogeneous atoms. Once you have selected this, you can colour them directly by right-clicking directly on the word: ‘col’ in the Control panel, and picking a strong colour. Then scroll down through the control panel, to find where the HETATM’s are. Create an image, like the one below, but which includes all four hetero atoms and DNA chains in the PDB file. Question 4. 1 What does this structural model represent? The model represents DNA recombination, a so called Holliday junction. 5. Analyzing interacting surfaces Obtain the PDB file 1dkf. It is a heterodimer complex of the ligand binding domains for RAR (retinoic acid receptor) and RXR (retinoid x receptor-alpha) nuclear receptors. Question: 5.1 Which chain corresponds to RAR and which to RXR? Hint: Look in the PDB file Chain A corresponds to RXR and chain B to RAR. 5.2 Which residues are responsible for the interaction between the two monomers? Start by finding intermolecular hydrogen bonds. Mark the residues that are hydrogen bonded to each other. Hint: Colour the backbones of the chains with different colours. Select one of the chains. Then Select: Groups Close to an other Chain …Select groups that are within 5 Å of another chain. This amounts to selecting the residues within 5 angstroms of the subunit interface. Press return to eliminate other residues. H-bonds: A-chain B-chain lys361, arg426 asp338 lys422 glu353 glu406 arg364 arg431 ser386 6. Comparing proteins Now we want to compare the single monomers in the RAR-RXR nucleic acid receptor heterodimer. From PDB import the files for 1exa (RAR monomer) and 1lbd (RXR monomer). Center and display the structures as ribbons. Colour each structure with a different colour (Hint: Color: by Layer. Remember to select ribbon from the control panel right most menu, before coloring). Find out how similar the two structures are by superimposing them on each other and checking from the structural alignment how many residue pairs are similar. Try first Fit: Magic Fit. Select 1exa as reference structure (the first selected is the reference) and accept default settings (“Use CA (carbon alpha) only”. Then Fit: Calculate RMS. Accept the default settings again. Then try improving the fit: Improved Fit and calculate RMS again. Question: 6.1 What is the RMSD (Root Mean Square Deviation) after doing Magic Fit of the two structures and what is the number of atoms used? What RMS do you get after improving the fit? What is the number of CA atoms used here? After Magic Fit RMSD is 6.82 Å (approximately). 189 CA atoms used. Look at the alignment window below. Scroll across it. The pairs that are highlighted are the residues which were aligned with each other by the fitting operation. By placing the pointer (without clicking) on a residue of the reference layer (the upper sequence), its identity appears on the left in the window and you will se it turn orange in the graphics window (if side chains are shown!). If you place the pointer on the corresponding residue in the lower sequence, you will see the distance between them. Question: 6.2 What is the number of identical amino acid pairs in the structural alignment? What is the number of similar amino acid pairs? What is the total number of residues in 1exa and 1lbd? Hint: Select: aa Identical to Ref. Structure. Look in Layers Infos window where the number of selected residues marked. Then Select: aa Similar to Ref. Structure. One way to get the total number of residues in a model is selecting first Select: Group Kind HETATM and then Select: Inverse Selection. Answer: There are 61 identical amino acid pairs and 123 similar amino acid pairs in the structural alignment. The total number of residues is 236 in 1exa and 239. 7. Displaying hydrogen bonds From molecule 1qmg, pick a single helix and display it in CPK format with hydrogen bonds. You should see something like the image below. You can switch on the hydrogen bond distances in the layers window. Question 7.1 What is the approximate distance of the hydrogen bonds (use the layers info, and then estimate the mean value yourself) Approximate distance is: 3 angstroms. For the same molecule, display all the sheets (or strands) in the molecule, and make a rough estimate of the distance of the hydrogen bonds (using Select:Secondary structure will help). Question: 7.2 Is there any difference in the distances of hydrogen bonds for helices and sheets? Very difficult to tell. The average hydrogen bond distance for all the strands could be anywhere between 2.8 and 3.0 (very roughly). It looks like the H-bond distance for sheets is either equal to or slightly smaller than the H-bond distance for helices.