Cristian_sup
advertisement

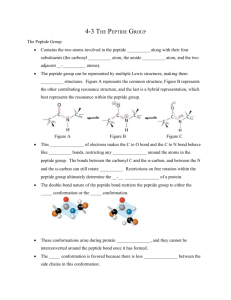
DETERMINATION OF MEMBRANE PROTEIN STABILITY VIA THERMODYNAMIC COUPLING OF FOLDING TO THIOL-DISULFIDE INTERCHANGE Electronic Supplementary Material Lidia Cristian, James D. Lear*, William F. DeGrado* Department of Biochemistry & Biophysics, University of Pennsylvania School of Medicine, Philadelphia PA, 19104-6059 U.S.A. Running title: Determination of Membrane Protein Stability * Authors to whom correspondences should be addressed. Addresses: Department of Biochemistry & Biophysics University of Pennsylvania School of Medicine 1010 Stellar-Chance Building Philadelphia PA, 19104-6059 U.S.A. Phone: (215) 898-4590 Fax: (215) 573-7229 Email: wdegrado@mail.med.upenn.edu The electronic supplementary material consists of 2 pages describing the procedure utilized to fit the data and the legends for two supplementary figures. File prepared using Microsoft Word 2000 from a PC. 1 The model presented in Scheme 2 was utilized to derive the equation for fitting the data in Figure 5. For fitting model parameters to the experimental data, the equilibrium concentrations of all species present were expressed as a function of known peptide and detergent concentrations using the equilibria in Scheme 2. The material balance in Scheme 2 can be expressed as: Equation 1 MSH + 4 TSH, SH + 4 T SS,SH + 4 T SS,SS + 2 DSS = Pt where Pt is the total protein concentration. MSH, TSH, SH, TSS,SH, TSS,SS and DSS concentrations, calculated as functions of MSH from the corresponding equilibrium constants defined in the text, were substituted in Equation 1 to give: Equation 2 MSH + 4MSH4/K1+4xMSH4/(K1K2)+4x2MSH4/(K1K2K3)+2[MSH4x2K4/(K1K2K3)]0.5–Pt = 0 where x represents [GSSG]/[GSH]2. This equation, solved using iterative root-finding algorithms in Igor Pro was used to individually fit data sets at fixed x in Figure 5. All parameters were allowed to freely vary. As initial guesses for the curve fit, we used K 1 and K4 values obtained from the equilibrium sedimentation study. K2 and K3 were given close starting values; generally K3 was chosen ~ (1/3) K2, as discussed in the text. The parameters fitted to the data are shown in Table 1. In fitting the data, we found it necessary to include in the fitting function a baseline correction factor, g. This parameter was also allowed to vary in all fits. This parameter presumably reflects the presence of some fraction of the peptide existing as disulfide-bonded species, which is insensitive to the redox potential of solution (probably due to the tendency of the peptide to form non- 2 specific aggregates). The values obtained for g were: 20.6, 42.1 and 39.2 corresponding to the data sets for 0.1, 0.2 and 0.5 GSSG/GSH ratios, respectively (see Figure 5). 3 Supplementary Figures Figure S1. Equilibrium analytical ultracentrifugation results for reduced (a) and oxidized (b) peptide in 15 mM DPC at 25 0C. The peptide-to-DPC ratio was 1:150 in both cases. The lines are best fits to a monomer-tetramer equilibrium model (a) and dimer-tetramer association scheme (b). The residuals of the fits are shown in the upper panels. 4 Figure S2. Plot showing the reversibility of the disulfide-bonding in DPC micelles. The circles represent the data when the equilibration is carried out starting with the reduced peptide. Diamonds correspond to data obtained in the reverse measurements when the equilibrium is approached from the other direction by starting with the dimer. The ratio of peptide/detergent was 1:50 in both experiments. 5