Methods Supplement
advertisement

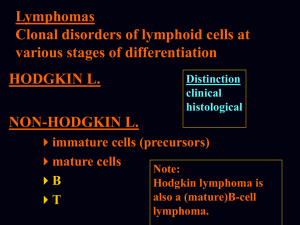
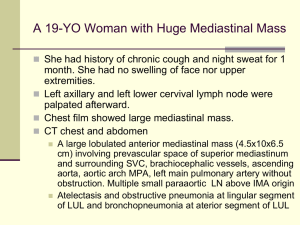
Ngo et al. Supplementary Methods Ref#: 2005-12-13892A Culture of lymphoma cell lines Lymphoma cell lines were cultured in RPMI, 10% FBS, except for OCI-Ly3, OCI-Ly10, OCI-Ly7, and OCI-Ly19, which were cultured in Iscove's modified Dulbecco medium with 20% human plasma. The OCI series of cell lines1 was provided by Hans Messner. The MedB1 cell line2 was provided by Peter Moller, the K1106 cell line3 was provided by Karen Leroy, the HBL1 cell line4 was provided by Martin Dyer and the U29405 and U29326 cell lines were provided by Rose-Marie Amini. Preparation of doxycycline-inducible cell lines Each cell line used in the bar code screen was first transduced with a feline endogenous virus (FEV) expressing the ecotropic retroviral receptor using bleomycin as the selectable marker. The FEV was constitutively produced in the supernatant of the producer line FLYRD187. This ecotropic receptor-expressing cell population was secondarily infected with a retrovirus expressing the bacterial tetracycline repressor (TETR) using hygromycin or neomycin as the selectable marker. The TETR was amplified by PCR from the pcDNA6/TR plasmid (Invitrogen) and cloned into the retroviral vector pBMNIRES (kindly provided by Gary Nolan). Single cell clones were screened for TETR expression by Western blot and for the ability of doxycycline to regulate expression of Photinus luciferase or GFP from an inducible retroviral vector in which the CMV promoter used to drive expression of the reporter contains TETR binding sites. Retroviral infection A mixture of shRNA constructs and the mutant ecotropic envelope-expressing plasmid pHIT/EA6x3*8 was used to transfect the CEB producer cells9 using the Lipofectamine 2000 reagent (Invitrogen). The retroviral library pool was used to infect doxycyclineinducible lymphoma cells in the presence of 8 g/ml polybrene in a single spin infection as described10 and puromycin was used to select for stable integrants over 6 days. Barcode DNA microarrays Bar code sequences were amplified from genomic DNA using the primers, 5’ CTAATACGACTCACTATAGGGAGATGTCGCTATGTGTTCTGGGAAATCAC 3’ and 5’ GGTTAAGATCAAGGTCTTTTCACCTGGC 3’. Amplified bar code sequences were in vitro transcribed using the MAXIscript T7 kit (Ambion) and the transcribed RNA products were labeled fluorescently with either Cy5 (induced) or Cy3 (uninduced) using the Universal Linkage System (Amersham Biosciences) as described11. Microarrays of bar code sequences were printed essentially as described12 using bar code oligonucleotides at 25 M in 3X SSC. Two micrograms of each labeled probe from induced and uninduced samples were combined and hybridized to DNA microarrays in 40% formamide, 5X SSC, 0.2% SDS, and 0.1 g/ul COT-1 DNA at 55 0C for 16 hours, which were then washed, scanned, and analyzed as described12. Each bar code experiment was performed in quadruplicate, and the microarray results for each bar code were averaged. Cell-based IKK assay Lymphoma cells expressing an IkB-Photinus luciferase fusion protein were used to measure IKK activity, as described10. IKK reporter cell lines were created from lymphoma cell lines expressing the ecotropic retroviral receptor and the TETR by sequential transduction wi -Photinus fusion protein10 and Renilla luciferases. Co-expressed Renilla luciferase was used to normalize for viable cell number. IKK reporter values were assayed over time using the Dual-Glo dual luciferase assay system (Promega). Gene expression profiling mRNA from cells induced to express CARD11 shRNA #1 was labeled fluorescently with Cy5 dye and mRNA from parallel uninduced cells was labeled with Cy3 dye, and these probes were co-hybridized to Lymphochip DNA microarrays as described12. Cytokine measurement Cells were cultured in fresh medium at the time of doxycycline induction and the concentration of IL-6 in culture supernatants at subsequent time points was measured by enzyme-linked immunosorbent assay (R&D Systems). IL-6 data were normalized to live cell number, and are representative of three independent experiments. Survival assay The puromycin-GFP fusion protein cassette was previously described13 and kindly provided by Enzo Medico. References 1. 2. 3. 4. 5. 6. 7. 8. 9. 10. 11. 12. 13. Tweeddale, M. E. et al. The presence of clonogenic cells in high-grade malignant lymphoma: a prognostic factor. Blood 69, 1307-14 (1987). Bentz, M. et al. Gain of chromosome arm 9p is characteristic of primary mediastinal B-cell lymphoma (MBL): comprehensive molecular cytogenetic analysis and presentation of a novel MBL cell line. Genes Chromosomes Cancer 30, 393-401 (2001). Copie-Bergman, C. et al. Interleukin 4-induced gene 1 is activated in primary mediastinal large B-cell lymphoma. Blood 101, 2756-61 (2003). Abe, M., Nozawa, Y., Wakasa, H., Ohno, H. & Fukuhara, S. Characterization and comparison of two newly established Epstein-Barr virus-negative lymphoma Bcell lines. Surface markers, growth characteristics, cytogenetics, and transplantability. Cancer 61, 483-90 (1988). Sambade, C. et al. U-2940, a human B-cell line derived from a diffuse large cell lymphoma sequential to Hodgkin lymphoma. Int J Cancer (2005). Amini, R. M. et al. A novel B-cell line (U-2932) established from a patient with diffuse large B-cell lymphoma following Hodgkin lymphoma. Leuk Lymphoma 43, 2179-89 (2002). Cosset, F. L., Takeuchi, Y., Battini, J. L., Weiss, R. A. & Collins, M. K. Hightiter packaging cells producing recombinant retroviruses resistant to human serum. J Virol 69, 7430-6 (1995). O'Reilly, L. & Roth, M. J. G541R within the 4070A TM protein regulates fusion in murine leukemia viruses. J Virol 77, 12011-21 (2003). Bupp, K. & Roth, M. J. Altering retroviral tropism using a random-display envelope library. Mol Ther 5, 329-35 (2002). Lam, L. T. et al. Small Molecule Inhibitors of IkB-Kinase are Selectively Toxic for Subgroups of Diffuse Large B Cell Lymphoma Defined by Gene Expression Profiling. Clin Cancer Res 11, 28-40 (2005). Berns, K. et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature 428, 431-7 (2004). Alizadeh, A. A. et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature 403, 503-511 (2000). De Palma, M. et al. Promoter trapping reveals significant differences in integration site selection between MLV and HIV vectors in primary hematopoietic cells. Blood 105, 2307-15 (2005).