(or Fluorescence) Resonance Energy Transfer (FRET)
advertisement
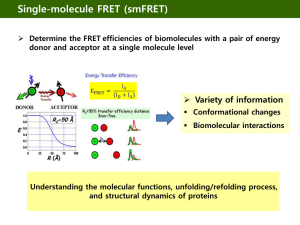
Resonance Energy Transfer Non-Radiative Energy Transfer driven by dipole-dipole coupling Fluorescence Resonance Energy Transfer Surface Energy Transfer Multiplicity : A property of a system due to the spin, or angular momentum, of its component particles ( e.g., electrons) Multiplicity is the quantification of the amount of unpaired electron spin - Hund's rule : favors the single filling of degenerate (same energy) Number of states with a given angular momentum : 2 S + 1, S= total spin if all electrons are paired, S=0: multiplicity =1; singlet if one unpaired electron, S = 1/2 ; doublet if two unpaired electrons, S =1 ; triplet Energy scheme used to explain the difference between fluorescence and phosphorescence Singlet state Triplet state Phosphorescence is a process in which energy absorbed by a substance is released relatively slowly in the form of light Fluorescence One of a class of luminescence phenomena in which certain molecules may emit light with a longer wavelength than the light with which were excited D + hv E D* ki D kf D + hv F Quantum yield Q The ratio of the number of photons emitted to the number of photons absorbed Lifetime The average time spent in the excited state before returning to the ground state Förster (or Fluorescence) Resonance Energy Transfer (FRET) Non-radiative energy transfer from an energy donor to an energy acceptor Dipole – dipole coupling Energy transfer efficiency : - Degree of spectral overlap between donor fluorescence emission and acceptor absorption - Inversely proportional to 6th power of the distance between fluorophores - ~ 10 nm Energy Level Diagram Use of FRET measurements Calculation of the distance between fluorophores Detection of target analytes Analysis of biomolecular interactions Single molecule analysis - Protein folding/unfolding - Protein dynamics Förster distance The relationship between the transfer efficiency and the distance between the two probe (R) Ro : the Förster distance at which the energy transfer is (on average) 50% Ro can be calculated using Qd : the quantum yield of the donor, n : the refractive index of the medium (generally assumed to be 1.4 for proteins) Nav : Avogadro's number (Nav= 6.02 x 10 per mole) Kappa squared : the orientation factor J : the overlap integral Kappa squared Orientation factor , kappa squared The overlap intergral J The degree of overlap between the donor fluorescence spectrum and the acceptor absorption spectrum λ : the wavelength of the light ε(λ) : the molar extinction coefficient of the acceptor at that wavelength f : the fluorescence spectrum of the donor normalized on the wavelength scale Surface Energy Transfer Energy transfer from a dipole to a metallic surface Interaction of the electromagnetic field of the donor dipole with the nearly free conduction electrons of the accepting metal Surface energy transfer efficiency : KSET = (1/τD) ( do/d)4 Yun et al., JACS, 2005, 127, 3115-3119 Schematic representation of the system, which consists of a fluorescein moiety (FAM) appended to ds-DNA of length R (varying from 15 to 60bp) with a Au nanoparticle (d = 1.4 nm) appended to the other end. - The flexible C6 linker produces a cone of uncertainty ( R) for both moieties. Addition of EcoRI (methyltransferase) bends the dsDNA at the GAATTC site by 128o , producing a new effective distance R'. 15 20 30 60 bp bp bp bp ; ; ; ; 62 A 96.4 A 130.4 A 232.4 A 10 bases per turn 3.4 A per base Efficiency vs distance -Energy transfer efficiency plotted versus separation distance between FAM and Au(NM). -Filled circles (·) represent DNA lengths of 15bp, 20bp, 30bp, and 60bp. The measured efficiencies of these strands with the addition of M.EcoRI are represented by the open circles. - The dashed line is the theoretical FRET efficiency, while the solid line the theoretical SET efficiency Conditions -Overlapping of Donar emission and Acceptor Excitation spectrum. -FRET : Donor/Acceptor; <10nm. -SET : Donor/Metal : <20 nm -Spectrally distinct Applications Pairs (http://probes.invitrogen.com/resources/; - Biomolecular interaction study in vivo/vitro - Tracking biomolecualr conformational change -In vivo imaging/co-localization study - Drug discovery - Bio-sensing //microscopy.biorad.com) - Organic dye -ALEA-488/RHOD-2; FITC/RHOD-2; FITC/TRITC; GFP/RHOD-2 - Fluorescent protein -BFP/GFP; BFP/YFP; BFP/RFP; CFP/YFP - Nanocrystal -QD/QD; QD/gold Examples of available fluorescent dye and quencher families. Tetramethylrhodamine (TMR), carboxytetramethylrhodamine (TAMRA), and carboxy-X-rhodamine (ROX) are all rhodamine-based dyes. The most common D/A dye combinations: coumarin/fluorescein, fluorescein/rhodamine, and Cy3.5/Cy5. Popular dye/quencher combinations: rhodamine/Dabcyl and Cy3/QSY9. Major suppliers: -Molecular Probes (fluorescein, rhodamine, AlexaFluor, BODIPY Oregon Green, Texas Red, and QSY quenchers), -Amersham Biosciences (Cy dyes and Cy5Q/Cy7Q quenchers) - AnaSpec (HiLyte Fluors, QXL quenchers) - ATTO-TEC (ATTO dyes and quenchers - Biosearch Technologies (Black Hole). FITC=fluorescein isothiocyanate. Fluorephore materials used in bioanalytical FRET Organic materials - Available in reactive form from commercial sources : activated with N- hydroxysuccinimide (NHS) ester, maleimide, hydrazide, amine functionality Ex) Fluorescein dyes : very popular because of their high quantum yield, solubility, ease of bioconjugation. Excitation with a standard argon-ion laser (488 nm) High rate of photo-bleaching, pH sensitive, self-quenching - Alternatives : AlexaFluore, Cy family, BODIPY Inorganic materials Metal chelates, semiconductor nanocrystals Biological origins - Fluorescent proteins Structures of common UV/Vis fluorescent dyes. Typical substituents at the R position include CO2-, SO3-, OH, OCH3, CH3, and NO2; Rx marks the typical position of the bioconjugation linker. Target Reactive Group Comment thiol maleimide, iodoacetyl, piridyldisulfide[a] site-specific but requires a free cysteine on proteins primary amine succinimidyl esters (NHS), sulfonyl chlorides, iso(thio)cyanates, carbonyl azides[a] proteins may have many primary amines carboxyl carbonyldiimidazoles, carbodiimides[b] allows further coupling to amines hydroxyl carbonyldiimidazoles, periodate, disuccinimidyl carbonate[b] allows further coupling to amines carbohydrates periodate[b] oxidizes sugars to create reactive aldehydes, which couple to amines intracellular proteins FlAsH requires cloning intracellular proteins SNAP-tag/HaloTag requires cloning and commercial ligands intracellular proteins fluorescent proteins requires cloning and formation of a chimera Experimental methods Conventional filter FRET Apply filter/emission band configurations for donor, acceptor and FRET (donor excitation and acceptor emission) to acquire single images or time series. If the donor signal decreases, acceptor and FRET signal increases. Acceptor photobleaching Apply donor/ acceptor configurations to acquire single images or time series. After some control images, acceptor (with 514 nm) is bleached. Donor signal increases after acceptor bleach Analysis of FRET Fluorescence lifetime imaging microscopy (FLIM) Information about the interactions between, and the structural states of, signaling molecules needs to be obtained as a function of space and time in a living cell. By using FLIM, the nanosecond decay kinetics of the electronic excited-state of fluorophores can be mapped spatially. Fluorescence lifetime The average amount of time that a molecule spends in the excited state upon absorption of a photon of light. Fluorescence lifetime is independent of fluorophore concentration and light-path length. Time domain FLIM Fluorescent proteins Green Fluorescence Protein (GFP) from jellyfish Widespread use by their expression in other organisms Key internal residues are modified during maturation to form the p-hydroxybenzylideneimidazolinon chromophore, located in the central helix and surrounded by 11 ß-strands (ß-can structure) In-vivo labeling of cells ; Localization and tracing of target protein GFP variants : BFP, CFP, YFP Red fluorescent protein (DS Red) from coral reef : tetrameric, slow maturation Monomeric RFP by protein engineering Quantum yield : 0.17 (BFP) ~ 0.79 (GFP) BFP/CFP ; CFP/YFP( high change in the FRET signal ratio) : fused to N- or C terminus of proteins by gene manipulation GFP (Green Fluorescent Protein) Jellyfish Aequorea victoria A tightly packed -can (11 sheets) enclosing an -helix containing the chromophore 238 amino acids Chromophore Cyclic tripeptide derived from Ser-Tyr-Gly The wt GFP absorbs UV and blue light (395nm and 470nm) and emits green light (maximally at 509nm) a) Normalized absorption and b) fluorescence profiles of representative fluorescent proteins: cyan fluorescent protein (cyan), GFP, Zs Green, yellow fluorescent protein (YFP), and three variants of red fluorescent protein (DS Red2, AS Red2, HC Red). From Clontech. Analysis of biomolecular interactions using FP Inter-molecular FRET FRET-based Sensors Intra-molecular FRET Calmodulin - Calcium ions : crucial for the metabolism and physiology of eukaryotes - - Regulate many cellular processes, ranging from transcription control and cell survival to neurotransmitter release and muscle function. Calmodulin (CaM, 148 aa); a ubiquitous, calciumbinding protein ( typically binds 0, 2 or 4 Ca+2) Regulate a multitude of different protein targets, affecting many different cellular functions. - CaM : mediates processes such as inflammation, metabolism, apoptosis, muscle contraction, intracellular movement, short-term and long-term memory, nerve growth and the immune response. - In the absence of Ca+2, the two main helical domains have hydrophobic cores. On the binding of a calcium ion, conformational changes exposes hydrophobic regions which have the potential to act as docking regions for target proteins ( over 100 proteins including kinases, phosphatases etc.) In the absence of Ca+2 In the presence of Ca+2 Modified MBP fluorescent indicator. ECFP as donor was fused to the N terminus of MBP, and YFP as a FRET acceptor was fused to the C terminus. - H indicates the portion of protein functioning as a hinge between the two lobes of the MBP. - The central binding pocket of the MBP is located between the two lobes. - In the absence of maltose, the two FPs are at their maximum distance from each other and FRET is minimal. Upon binding maltose, the MBP undergoes a conformation change that brings the two FPs into close proximity and increases FRET, which can be monitored by the change in ratio of the YFP and CFP emission - a) Confocal image of a maltose-FP sensor expressed in yeast. Fluorescence is detected in the cytosol but not in the vacuole. Scale bar=1 um. b) Changes of the maltose concentration in the cytosol of yeast that expresses a maltose sensor with a Kd value of 25 nM. The graph indicates emission ratio as a function of maltose uptake for a single yeast cell. Enzyme-generated Bioluminescence - BRET ( Bioluminescence RET) : Donor : Luciferase ; Acceptor : GFP - No excitation light source to excite the donor, which avoids problems such as light scattering, high background noise, and direct acceptor excitation In-vivo monitoring of protein-protein interactions such as circadian clock proteins, insulin receptor activity, real-time monitoring of intracellular ubiquitination The firefly luciferase/luciferin system : the best candidate for a BRET-based donor ; high quantum yield ( 0.88) Bioluminescent substrates and enzymatic reactions of several common luciferases: a) the aliphatic aldehyde substrate of bacterial luciferase; b) structure and reaction of luciferin, the substrate of firefly luciferase; c) colenterazine, the substrate for Renilla luciferase and also part of apoaequorin. Renilla (Sea pansy) Enzyme-generated Chemiluminescence : Luminophore : synthetic substrate that is excited through an enzymatically catalyzed reactions Chemiluminescent substrates and the enzymatic reactions of horseradish peroxidase (HRP) and alkaline phosphatase. a) Luminol; b) Acridan (also available as an ester); c) Adamantyl-1,2-dioxetane (substrate for alkaline phosphatase and other enzymes). Self-illuminating quantum dot conjugates for in-vivo imaging Unique optical property of Qd: - High quantum yields, large molar extinction coefficients, size-dependent tunable emission and high photostability Fluorescent probes for biological imaging Challenging issues - Requirement for external illumination strong background auto-fluorescence from ubiquitous endogenous chromophores such as collagen, porphyrins and flavins little light is available for quantum dot excitation at non-superficial locations due to absorption and scattering of optical photons in tissues Ideal quantum for in-vivo imaging - Light emission with no requirement for external excitation Quantum dot conjugates based on the principle of BRET So et al., Nature Biotech., 24, 339-343 (2006) Construction of self-illuminating QDs conjugates Use of luciferase from Renilla reniformis Luc8 : Eight mutation variant : more stable in serum and higher catalytic activity - emit blue light with a peak at 480 nm upon addition of its substrate coelenterazine Conjugation of Luc8 to polymer-coated CdSe/ZnS core-shell QD 655 through coupling of the amino groups on LUC8 to carboxylated on the QD - The hydrodynamic diameter of QD655-Luc8 : ~ 2 nm - The conjugate contains six copies of Luc8 on average Gold nanoparticles Exceptional quenching ability Plasmon resonances in the visible range with large extinction coefficient (105 /cm/M) Stable Unfluctuating signal intensities Resistant to photo-bleaching Gold Nano Particles (AuNPs) Core Materials for NPs - Au, Ag, Pt : Electron transporter, Catalysis, NPs coating for electrode - Mg, Co, Fe : Magnetic behavior, Sample purification, MRI signal enhancing - CdSe, ZnS, InP : Semiconductor QDs Stabilization by surfactants in synthesis of AuNPs - Reduction of HAuCl4 in the presence of surfactant - Citrate, tannic acid, white phosphorus : > 3 nm - Alkanethiol : Monolayer protected cluster (MPC), 2 ~ 3 nm - Dendrimer : Dendrimer encapsulated nanocluster (DEN). < 2 nm Characteristics of AuNPs - Surface Plasmon Resonance Band . Absorbance band near 520 nm in 5 ~ several tens nm of AuNPs . SPB shift responding to surface modification and environmental condition - Photoluminescence as Gold QDs . <2nm of AuNPs : smaller Bohr radius than semiconductor . Size dependent excitation/emission spectrum Synthesis of AuNPs NP : Nanoparticle capped with surfactant (ex) sodium citrate MPC : Monolayer-Protected Clusters with alkanethiol (ex) 1-OT / 11-MUA DEN / DSN : Dendrimer-Encapsulated (or Stabilized) Nanoclusters Reduction NaBH4 DSN Surfactant NP MPC Gold Nanoparticles Gold Quantum Dot before DEN reduction AuCl4- DSN (G2-NH2/OH) Citric Acid DEN (G4) MPC Molecular Beacon (MB) Single-stranded oligonucleotide molecular probe Target strand Quenching by FRET Donor : FAM (Ex 494 nm, Em 520 nm) Quencher : Dacyl ( Abs 380 ~600 nm) FRET-based probe : Sensitive and no separation step Real time analysis of amplicons by PCR Sequence specific multiplex analysis Molecular Beacons Single-stranded oligonucleotide hybridization probes that form a stem-and-loop structure The loop contains a probe sequence that is complementary arm sequences that are located on either side of nthe probe sequence A fluorophore is covalently linked to the end of one arm and a quencher is covalently linked to the end of the other arm The probe is dark in the absence of targets. When the probe encounters a target molecule, it forms a probe-target hybrid that is longer and more stable than the stem hybrid The molecular beacon undergoes a spontaneous conformational reorganization that leads the stem hybrid to dissociate and fluorophoree and the quencher to move away form each other, restoring fluorescence Use : genetic screening, SNP detection, pharmacogenetic applications Diagnosis of acquired resistance in lung cancer using molecular beacons Lung cancer One of the most common cancers : Increased rate of 8.7 % Leading cause of cancer death 5-year survival rate : 60 % at stage 1 30 % at stage 2 10 % at stage 3 Early diagnosis rate: < 25 % Key issue in lung cancer treatment Acquired drug resistance - Severe reduction in the drug efficacy during chemotherapy - Patients who initially respond develop acquired resistance to drug - Major cause of lung cancer death Acquired drug resistance in lung cancer Inhibitors against EGFR-TK domain : Gefitinib (IRESSA) and Erlotinib (Tarceva) Unknown Mechanisms Erlotinib T790M secondary mutation MET amplification Mutations in EGFR TK domain - Primary mutations : L858R, Exon 19 deletion Susceptible to TKIs - Secondary mutation: Thr to Met at codon 790 (T790M) Resistance to TKIs Diagnosis of acquired resistance: crucial for deciding a chemotherapy Direct sequencing : laborious and time-consuming Design of Molecular Beacon for T790M Secondary mutation : Thr to Met at codon 790 (T790M) of EGFR 790 C T CT Sequence(5`3`) Synthetic target sequence(5`3`; 60bp) WT-MB Cy3-TCGGAGCTGCGTGATGAGTCCGA-Dabcyl ACCTCCACCGTGCAGCTCATCACGCAGC TCATGCCCTTCGGCTGCCTCCT 2`MT-MB FAM-TCGGAGCTGCATGATGAGTCCGA-Dabcyl ACCTCCACCGTGCAGCTCATCATGCAGC TCATGCCCTTCGGCTGCCTCCT Control MB Cy3-CCAAGACGTTCAGATTCGCTTGG-Dabcyl Real (A)time PCR for detecting T790M (B) FAM-Mut1 Cell1line 2 350 only MB 300 W/O templ 250 PC9 (A)PC9 gDNA 200 Flu 3 4 EGFR genotype Exon19 del (B) H1975 L858R / T790M H1975 gDNA 150 FAM-Mut1 500 100 1 350 50 W/O templ 250 -50 Flu 10 200 150 3 4 only MB 300 0 2 100 PC9 gDNA 15 20 25 H1975 gDNA 100 30 35 500 cycle 50 Amplicon: 150 bp containing EGFR 2nd mutation 0 40 Cycles -50 100 10 at 95 15 °C, Annealing 20 25 - Denaturing at 3060 °C 35 cycle - Monitoring of fluorescence at the annealing step - Extension at 72 °C Oh et al., J Mol. Diag. (2011) •Lane1 : FAM-Mut MB + buffer •Lane2 : FAM Mut MB + PCR buffer without template •Lane3 : PCR with PC9 genomic DNA •Lane4 : PCR with H1975 genomic DNA Detection sensitivity for T790M Biopsy sample : a mixture of cancer and normal cells Relative ratio of 2nd mutated EGFR gene (T790M) to the wild type Cell line EGFR genotype PC9 Exon19 del H1975 L858R / T790M Detection sensitivity: ~ 2 % EGFR gene with T790M Direct sequencing : ~ 25 % of 2nd mutated EGFR gene Oh et al., J Mol. Diag. (2011) Test for real patient samples Tube No Sample Gefitinib treatment EGFR T790M Concentration 27P Primary lung cancer Post L858R Y 100 ng/μl 28M1 Contralateral lung Post L858R Y 100 ng/μl gDNA extraction from biopsy samples from lung cancer patients followed by real-time PCR Positive control : H1975 Negative control : PC9 gDNA template 100 ng 95 °C 30s 60 °C 30s PC9 28M1 1.00 Fluorescence (A.U.) Supplied by Mitsudomi group at Kyoto University 27P H1975 0.60 0.20 50 cycles -0.20 5 Oh et al., J Mol. Diag. (2011) 15 25 Cycles 35 45 Fluorescence imaging using molecular beacons More simple diagnosis of acquired resistance Fixation & permeabilization Incubation with MB for 40 min for 20 min Imaging by Confocal microscope H1975 2nd mutation PC9 targeted MB (T790M-MB) Donor : FAM (Ex. 494nm, Em 520nm) Quencher : Dabcyl dye (Absorption 380~ 600 nm) Drug Resistant Fluorescence imaging H1975 (T790M) FAM Cy5 Merge x60 FAM Cy5 Merge x60 PC9 T790M-specific MB Control MB : Cy5 : T790M MB– FAM stem-AA…AA-stem Analysis of biopsy samples with drug resistance by imaging Biopsy samples from cancer patients with drug resistance: 27 Detection of T790M by imaging using molecular beacons : 14 - Acquired resistance via other mechanisms: MET(Receptor tyrosine kinase) amplification Detection by conventional direct sequencing: 2 - Low sensitivity due to the normal cells in biopsy samples T790M positive (Patient # 38) T790M negative (Patient # 35) Proteolytic activity monitored by FRET between quantum dot and quencher QD–peptide sensor architecture and optical characteristics of the fluorophores used. (a) Schematic diagram of the self-assembled QD– peptide nanosensors; one peptide shown for clarity. Dyelabelled modular peptides containing appropriate cleavage sequences are self-assembled onto the QD. FRET from the QD to the proximal acceptors quenches the QD PL. Specific protease cleaves the peptide and alters FRET signature. (b) Normalized absorption and emission profile of dyes and QDs used: Cy3 dye (quantum yield=0.20, =150,000 M-1 cm-1, ex 555 nm, em 570 nm), QXL-520 dark dye quencher ( =26,000 M-1 cm-1, ex 508 and 530 nm). The absorption of the 538 nm QDs and the emission spectra of the 510 and 538 nm QDs are shown. (C) Model structure for the QD–peptide conjugates. Data were derived separately but both conformers are shown on the same QD. The Casp1–Cy3 peptide is shown on the right and the Thr–QXL on the left. A CdSe–ZnS core-shell QD with a diameter of 28–29 Å is represented by the blue inner sphere. For both peptides the His6 sequence shown in green is in contact with the QD surface in an energy minimized conformation. Protease recognition sequences are highlighted in yellow, and the spacer-linker sequences are shown in grey. The Cy3 acceptor dye structure is shown in red, and the QXL-520 quencher is approximated by a magenta sphere placed 10.5 Å from the cysteine S atom. The centre-to-centre distance determined from FRET efficiencies are 55 Å for the QD–Casp1–Cy3 (R0=54 Å) and 56 Å for QD–Thr– QXL (R0=43 Å). The second grey shell represents the DHLA ligand cap whose maximum lateral extension away from the QD surface can vary between 5 and 11 Å; 10 Å is shown here. Advantage vs shortcomings of FRET Advantage - Relatively cheap - Very efficient in measuring changes in very proximal distances - Measure distances in molecules in solution - Only need a few µM of labeled proteins - Rapid detection Shortcomings - Uncertainty of the orientated factor - When measuring a change in distance between two probes, the result is a scalar and give no indications of which probe (donor and/or acceptor) moves. - The presence of free labels in solution could mask a change in energy transfer. a) Structure of sulfo-NHS-BIPS (sulfo-NHS=N-hydroxysulfosuccinimide sodium salt; BIPS=1 ,3 ,3 -trimethylspiro[2H-1benzopyran-2,2 -indoline]) in the spiropyran (SP) form before (left) and merocyanine (MC) form after (right) conjugation to a protein. b) Schematic representation of quantum dot (QD) modulation by photochromic FRET after interacting with MBPBIPS (MBP=maltose-binding protein). When BIPS is converted to the MC form by UV light, the QD emission is reduced through FRET quenching. After photoconversion with white light to the SP form, the direct emission of the QD is substantially increased. c) Photoluminescence spectra of the 555-nm luminescing QD 20 MBP-BIPS system with a dye/protein ratio of 5 after photoconversion from the SP to the MC form. d) Effect of pcFRET on QD photoluminescence (initial change from white light to UV). Figure adapted from reference [106] with permission of the American Chemical Society.