Sodium Bisulfite Methods
advertisement
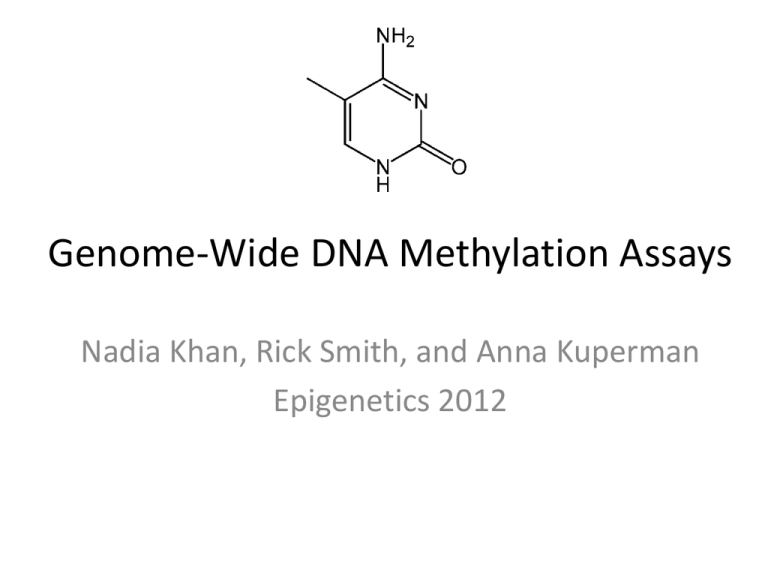
Genome-Wide DNA Methylation Assays Nadia Khan, Rick Smith, and Anna Kuperman Epigenetics 2012 Introduction • Most Genome Wide Approaches were adapted from technologies originally developed for detecting methylation at the level of a single gene • Advantages of a Genome Wide Approach – Scale of information • Whole chromosome • Whole genome – Wider regulatory networks – Facilitates comparative and population level analysis Tollefsbol 2009 Many methylation methods can be grouped into major categories based upon their general principles Genome-wide DNA Methylation Assays Nadia Rick Anna Methylation Sensitive Endonuclease- based Sodium Bisulfite Treatments Biological Affinity- based Methods 1. Restriction Landmark Genomic Scanning (RLGS) 1. Whole and targeted 1. Methylated DNA Immunoprecipitation (MeDIP) 2. Microarray coupling of Methylated-CpG island amplification 2. Bisulfite Libraries 2. Methylated CpG island recovery assay Concept of Methylation Sensitive Endonuclease Assays • Use a restriction enzyme(s) that is methylation specific and separate the unmethylated from the methylated • Unmethylated regions enzyme-sensitive Methylated regions enzyme-resistant • Identify multiple de novo methylated areas across genomes vs. one specific area – 2D gel and scintillation counting – Array Laird 2010 Restriction Landmark Genomic Scanning (RLGS) • Cleave genome into pieces based upon restriction landmarks (sites) • Radioactively label cleaved ends • Separate using 1D and 2D gel electrophoresis • Quantitate signal by amount of fluoresence in gel – Intensity = copy number of the restriction site Tollefsbol 2009 • NotI: • Radioactively labels ends • LEAST specific • EcoRV: • MORE specific • HinfI: • MOST specific Eng et al. 2000 RLGS in-use Takamiya, et al. 2009 Microarray Coupling of Methylated CpG Island Amplification (MCAM) • Cleave genomic DNA with SmaI (methylationsensitive) • Cut received pieces again with XmaI; create sticky ends • Amplify pieces using PCR • Hybridize onto a microarray • Analyze fluorescence reads and identify corresponding genomic address Tollefsbol 2009 Tollefsbol 2009 MCAM in-use Estecio 2007 Genome-wide DNA Methylation Assays Nadia Rick Anna Methylation Sensitive Endonuclease- based Sodium Bisulfite Treatments Biological Affinity- based Methods 1. Restriction Landmark Genomic Scanning (RLGS) 1. Whole and targeted 1. Methylated DNA Immunoprecipitation (MeDIP) 2. Microarray coupling of Methylated-CpG island amplification 2. Bisulfite Libraries 2. Methylated CpG island recovery assay Sodium Bisulfite Methods • Bisulfite Sequencing • Targeted and Whole Genome approaches • Bisulfite Libraries Bisulfite sequencing • m-C are resistant to bisulfite conversion • Compare with unconverted reference sequence to infer methylation pattern • Allows for single base resolution, but technically challenging genome wide Tollefsbol 2011 Whole Genome Approach • Pair Bisulfite Conversion with Next Generation Sequencing (NGS) – Massively parallel sequencing • Roche 454, Illumina, SOLiD platforms • Quick, relatively cheap, large scale analysis Tollefsbol 2011 Pyrosequencing Whole Genome Approach • Pair Bisulfite Conversion with Next Generation Sequencing (NGS) – Massively parallel sequencing • Roche 454, Illumina, SOLiD platforms • Quick, relatively cheap, large scale analysis • Tenable for relatively small genomes – Arabidopsis thaliana • Significant challenges for mammalian genomes – Reduced complexity of the genome – Short sequence reads – Solutions • Longer sequence reads • Targeted approaches Cokus et al. 2008; Tollefsbol 2011 Targeted Bisulfite Sequencing • Reduced Representation Bisulfite Sequencing (RRBS) • Molecular Inversion Probes (MIP) – Padlock Probes Targeted Bisulfite Sequencing • Reduced Representation Methylation Sequencing (RRMS) – Enrichment for CG-rich regions via Msp1 digestion (5‘-CCGG-3‘) – NGS – Disadvantages: mostly un-methylated DNA Meissner 2005; Jeddeloh 2008 Targeted Bisulfite Sequencing Padlock Probes Deng et al. 2009 Bisulfite Libraries • Applications and Advantages • • • • • Coverage of relevant genome regions Facilitates large comparative study Multiplex Sequencing High sensitivity Whole library amplification • Pair with NGS or Array Gu et al. 2011 Genome-wide DNA Methylation Assays Nadia Rick Anna Methylation Sensitive Endonuclease- based Sodium Bisulfite Treatments Biological Affinity- based Methods 1. Restriction Landmark Genomic Scanning (RLGS) 1. Whole and targeted 1. Methylated DNA Immunoprecipitation (MeDIP) 2. Microarray coupling of Methylated-CpG island amplification 2. Bisulfite Libraries 2. Methylated CpG island recovery assay Biological Affinity Based Methods • Basic Concept: Use antibodies that are specific for 5meC or proteins that bind preferably to methylated genomic DNA to profile DNA methylation patterns. These patterns are detected through microarrays or parallel high through-put sequencing. • Types – MBD affinity column (MAC) – Methylated DNA Immunoprecipitation (MeDIP) – Methylated-CpG island recovery assay Laird 2010 MBD Affinity Column (MAC) • Uses MeCP2, a member of a family of proteins that contain methyl-CpG binding domains. • Developed by Cross et al in 1994. This is the first time affinity enrichment was used to look at genome-wide methylation Methylated CpG Unmethylated CpG Figure 9.8 Tollefsbol 2009 MBD Affinity Column (MAC) • Advantages: this method is fast and efficient. • Limitations: Needs a large amount of starting genomic DNA to pass through column purification Figure 9.8 Tollefsbol 2009 Methylated DNA Immunoprecipitation (MeDIP) • Introduced in 2005 by Weber et al • Uses an antibody that specifically binds to methylated cytosines. • Fragmented DNA is incubated with the antibodies, immunoprecipitated, and then enrichment is quantified. • Advantages: Efficient, sensitive, large-scale analysis of genomic methylation • Limitations: need good quality 5meC antibodies and denatured ssDNA, which can be difficult to obtain in CpG rich genes, is required for analysis Figure 9.8 Tollefsbol 2009 Methylated DNA Immunoprecipitation (MeDIP) Methylated CpG Unmethylated CpG Figure 9.7 Tollefsbol 2009 MeDIP • How can it be used? – Identifying genes involved in cancer development • Ex: Morris et al were able to shortlist genes involved in renal cell carcinoma (RCC) suppression by looking at promoter regions that were frequently methylated in RCC lines, but not in normal kidney cell lines. Morris et al. 2011 Methylated-CpG Island Recovery Assay (MIRA) • MBD2b/MBD3L1 complex has a high affinity to methylated DNA (higher than MBD2b on it’s own; MBD3L1 has no affinity) • MIRA developed in 2006 by Rauch et al to have a better screen for methylation patterns in lung cancer tumors so can have a better early detection • Advantages: Can be used to examine large number of genes simultaneously, works on dsDNA, only need a few hundred nanograms of genomic DNA Figure 9.7 Tollefsbol 2009 Methylated-CpG Island Recovery Assay (MIRA) Figure 9.9 Tollefsbol 2009 From activemotif.com MIRA • Rauch et al (2006) were able to identify lung tumor suppressor genes • Rauch et al (2009) were able to use MIRA to characterize a human B-cell methylome at 100 bp resolution Rauch 2006 • A lot of these assays are commercially available • MeDIP – MagMeDIP Kit TM (Diagenode), – Methylated-DNA IP Kit (Zymo Research) and Methylamp™ – Methylated DNA Capture Kit (Epigentek) • MIRA – Ex: MethylCollector TM Ultra Biological Affinity Assays • Why are they good? – Quick and efficient genome-wide assessment of DNA methylation • Disadvantages: – Do not give information on individual CpG dinucleotides – Require experimental or bioniformatic adjustment for changing CpG density at different regions of genome Laird 2010 Complications with 5hydroxymethylcytosine • 5-hydroxymethylcytosine has been discovered in mammalian DNA, and is produced by an enzymatic pathway involving TET1 hydroxylase. • All 3 methods discussed are unable to distinguish between 5mC and 5hmC. • However, one can distinguish 5-hmc by adding a glucose to the hydroxy-group (EpiMark Kit) Tollefsbol 2009 The Future of Genome-Wide Methylation Assays • As more data is experimentally collected about the methylome, there will be more and more need for analysis. Bioinformatics is beginning to play a big role. • Increasing role of sequencing as opposed to arrays. • Nanopore sequencing could directly allow sequencing of 5meC References Cross SH et al. 1994. Purification of CpG islands using a methylated DNA binding column. Nat. Genet. 6(3):236-44. Deng J et al. 2009. Targeted bisulfite sequencing reveals changes in DNA methylation associated with nuclear reprogramming. Nat Biotechnol. 27(4):341-2. Estecio MRH et al. 2007. High-throughput Methylation Profiling by MCA Coupled to CpG Island Microarray. Genome Research 17(10): 1529-536. Gu H et al. 2011. Preparation of reduced representation bisulfite sequencing libraries for genome-scale DNA methylation profiling. Nature Protocols 6:468–481. Jeddeloh JA et al. 2008. Reduced-representation methylation mapping. Genome Biology 9:231. Laird PW. 2012. Principles and Challenges of Genome-wide DNA Methylation Analysis. Nature Reviews Genetics 11:191-203. Meissner A et al. 2005. Reduced representation bisulfite sequencing for comparative high-resolution DNA methylation analysis. Nuc. Acids Res. 33(18):5868-5877. Morris MR et al. 2011. 6.Genome-wide methylation analysis identifies epigenetically inactivated candidate tumour suppressor genes in renal cell carcinoma. Oncogene 30(12):1390-401. Rauch TA and Pfeifer GP. 2009. Chapter 9: Methods for Assessing Genome Wide DNA methylation. In: Handbook of Epigenetics : The New Molecular and Medical Genetics. ed. Tollefsbol T. Academic Press. Rauch TA et al. 2006. MIRA-assisted microarray analysis, a new technology for the determination of DNA methylation patterns, identifies frequent methylation of homeodomain-containing genes in lung cancer cells. Cancer Res. 66(16)793947. Rauch TA et al. 2009. A human B cell methylome at 100 base pair resolution. Proc. Natl. Acad. Sci. 106(3):671-8. Takamiya et al. 2009. The Application of Restriction Landmark Genome Scanning Method for Surveillance of NonMendelian Inheritance in F1 Hybrids. Comparative and Functional Genomics 2009: 1-7. Tost J. 2009. Epigenetics. Caister Academic Press. Weber M et al. 2005. Chromosome-wide and promoter-specific analyses identify sites of differential DNA methylation in normal and transformed human cells. Nat Genet. 37(8):853-62.