RNA Sequencing - Bioinformatics Institute
advertisement

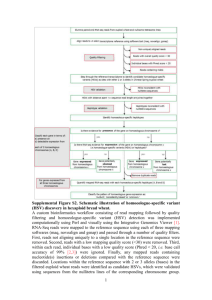
RNA Sequencing Peter Tsai Bioinformatics Institute, University of Auckland What is RNA-seq? Study of transcriptomes Identify known genes, exons, splicing events, ncRNA, miRNA Novel genes or transcripts Abundances of transcripts (quantitive expression) Differential expressed transcripts between different conditions Reconstructing transcriptome. General workflow Raw data QC Map to reference genome De novo transcriptome assembly Estimate abundance Normalisation Differential expression analysis Require downstream annotation Quality checks and mapping Use FastQC, SolexQA Trim off low quality region, keep only proper-paired reads Most QC software assume normality, but in RNA-seq data you will probably see none-normality You might see some duplicated reads, its probably due to highly expressed gene. Specific reference mapping tool that can map across splice junctions between exons, i.e. Tophat Specific de novo transcriptome assembly software for reconstruction of transcriptomes from RNA-seq data, i.e. Trinity Expression value in RNA-seq The total number of reads mapped to a gene/transcript (Count data or raw counts or digital gene expression) Complexity of using simple counts Sequencing depth: the higher the sequencing depth, the higher the counts Gene length: Counts are proportional to the length of the gene times mRNA expression level Counts distribution: difference on how counts are distributed among samples. Normalisation RPKM (Mortazavi et al, 2008) ◦ Reads Per Kilobase of exon model per Million mapped reads FPKM (Mortazavi et al, 2010) ◦ Fragments Per Kilobase of exon model per Million mapped reads ◦ Paired-end RNA-Seq experiments produce two reads per fragment, but that doesn't necessarily mean that both reads will be mappable. Replicate 2 Data exploration Replicate 1 Gene.ID/Description 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 logFC logCPM LR 2.563086301 5.07961611 4.003686266 2.330395704 2.71372512 9.704651395 -2.052703196 3.402621025 1.95117636 4.438847349 2.465833373 12.20593577 1.817858683 5.308092036 1.577603322 6.556675456 1.20515812 4.542565518 1.233090336 10.08249873 1.120581944 12.14988136 1.045292369 4.913492018 1.089867189 3.885246135 1.353955354 2.21406615 1.049933686 3.281031472 -1.032999983 1.480514873 -1.313778857 4.325330722 0.864451602 4.338668381 -0.766266641 5.2972332 PValue FDR 28.4599795 9.57E-08 2.72E-05 28.3288251 1.02E-07 2.72E-05 25.01930526 5.68E-07 0.000100653 21.11492168 4.33E-06 0.000575287 19.21195535 1.17E-05 0.001244651 10.91756889 0.000952565 0.084460792 10.3738524 0.001278126 0.097137553 9.690419768 0.001852312 0.110687766 9.670466698 0.001872537 0.110687766 9.289827985 0.002304298 0.122588652 7.710102379 0.005491264 0.265577482 7.039209923 0.00797442 0.350270537 6.912558621 0.008559242 0.350270537 5.976193603 0.014500264 0.551010036 5.737563572 0.016605812 0.588952795 4.712476717 0.029944481 0.995653998 4.169234925 0.041164384 0.998742102 3.479808135 0.062121942 0.998742102 3.443865378 0.063486998 0.998742102 Up-regulated Down-regulated ERCC spike-in control Set of external RNA transcripts with known concentration. Dynamic range and lower limit of detection Fold-change response Internal control, in order to measure against defined performance criteria Dynamic range and lower limit of detection The dynamic range can be measured as the difference between the highest and lowest concentration. Measure of sensitivity, and it is defined as the lowest molar amount of ERCC transcript detected in each sample Fold-change response How much library depth is needed for RNA-seq? Depends on a number of factors ◦ Biological questions Complexity of the organism Types of analysis Types of RNA, miRNA, lncRNA. Literature search for similar work Pilot experiment Summary Have 3 or more biological replicates Analysis your data with different normalisation methods Perform data exploration Use a standard spike-in as internal control Validation with qPCR