Lee Katz - Compgenomics2014
advertisement

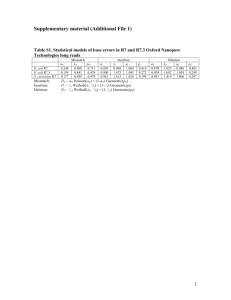
Computational assembly for prokaryotic sequencing projects Lee Katz, Ph.D. Bioinformatician, Enteric Diseases Laboratory Branch January 15, 2014 Disclaimers The findings and conclusions in this presentation have not been formally disseminated by the Centers for Disease Control and Prevention and should not be construed to represent any agency determination or policy. The findings and conclusions in this [report/presentation] are those of the author(s) and do not necessarily represent the official position of CDC Partners in Public Health Graduated Oct 2010 CDC 2010 - present Lee Katz, Present • Currently in the National Enteric Reference Laboratory • Vibrio, Campylobacter, Escherichia, Shigella, Yersinia, Salmonella • Focusing on Listeria and Vibrio One of my projects is #2 on CDC’s list of accomplishments for 2013! #2 http://www.cdc.gov/features/endofyear/ Outline • Sequencing – 1st gen – 2nd gen – 3rd gen • Reads – Quality control (Q/C) • Read metrics – Read-cleaning • Assembly – Algorithms – Assembly metrics Prokaryotic Sequencing Projects Stages • Sequencing • Assembly • Feature prediction • Functional annotation • …analysis… • Display (Genome Browser) Sequencing Assembly Examples • Haemophilus influenzae • Neisseria meningitidis • Bordetella bronchisceptica • Vibrio cholerae • Listeria monocytogenes prediction annotation Display Fleischman et al. (1995) “Whole-Genome Random Sequencing and Assembly of Haemophilus influenzae Rd” Science 269:5223 Kislyuk et al. (2010) “A computational genomics pipeline for prokaryotic sequencing projects” Bioinformatics 26:15 Out with the old; in with the new: Two new technologies to the compgenomics class! • • • • 454 Illumina single end reads Illumina paired end reads PacBio Sanger Sequencing (1st gen) Sequencing: first generation Sheer DNA Margulies et al. (2005) Genome sequencing in open microfabricated high density picoliter reactors. Nature 437:7057 Cloning into bacterial vectors Amplification Sanger sequencing Sanger sequencing output • Usually .ab1/.scf file format 454 Sequencing (2nd Gen) A 454 Pyrosequencing + PCR Reagents + Emulsion Oil B Mix DNA library & capture beads (limited dilution) “Break micro-reactors” Isolate DNA containing beads Create “Water-in-oil” emulsion Perform emulsion PCR 454 Pyrosequencing Load enzyme beads Load beads into PicoTiter™Plate PicoTiter™Plate Diameter = 44 μm 44 μm Depth = 55 μm Well size = 75 pl Well density = 480 wells mm-2 1.6 million wells per slide 454 Pyrosequencing Sequencing by synthesis Photons generated are captured by CCD camera Reagent flow Margulies et al., 2005 454 sequencing output Flowgram (.sff file format) Flow Order 4-mer 3-mer 2-mer 1-mer T A C G KEY (TCAG) Measures the presence or absence of each nucleotide at any given position Illumina sequencing (2nd Gen) The following animations are courtesy of Illumina, Inc. Region complementary to P5 grafting primer Index 2 P5 primer DNA insert P7 primer Index 1 P5 grafting primer P7 grafting primer Flow cell surface The following animations are courtesy of Illumina, Inc. SBS Sequencing Primer Hybridization The following animations are courtesy of Illumina, Inc. Sequence (Cycle 1) Sequence (Cycle 1) Index 1 Seq Primer Hybridization Index 1 read – 8 cycles Unblock P5 grafting primer 7 dark cycles P5 grafting primer Index 2 index read 8 cycles 7 dark cycles P5 grafting primer Index 2 index read 8 cycles 7 dark cycles P5 grafting primer Linearization Original strand New strand Illumina sequencing video • http://www.youtube.com/watch?v=womKfik WlxM PacBio sequencing* (3rd Gen) *Pacific Biosciences http://www.youtube.com/watch?v=NHCJ8PtYCFc SMRT Bell Zero-mode waveguide (ZMW), a very fancy and very small well Thanks to PacBio for donating some slide materials in this section Eid et al Science, January 2009/10.1126/science.1162986 http://www.youtube.com/watch?v=NHCJ8PtYCFc Eid et al Science, January 2009/10.1126/science.1162986 Eid et al Science, January 2009/10.1126/science.1162986 PacBio video http://www.youtube.com/watch?v=NHCJ8PtYCFc Q/C + cleaning + metrics READS Q/C • You need to know if your data are good! • Example software – FastQC – Computational Genomics Pipeline (CG-Pipeline) Quality Control FastQC output Quality Control bioinformatics FastQC output The CG-Pipeline way run_assembly_readMetrics.pl File tmp.fastq avgReadLength 80.00 totalBases 177777760 minReadLength 80 maxReadLength 80 avgQuality 35.39 Read cleaning Read cleaning with CG-Pipeline (not validated; please use with caution) F. Read R. Read Read %ACGT Phred http://sourceforge.net/projects/cg-pipeline/ Graphs made with FastqQC (AMOS) 1. Trimming low-qual ends run_assembly_trimLowQualEnds.pl F. Read R. Read Read 1A. %ACGT 1B. Phred http://sourceforge.net/projects/cg-pipeline/ Graphs made with FastqQC (AMOS) 2a. Removing duplicate reads 2b. Sometimes: downsampling run_assembly_removeDuplicateReads.pl Trimmed reads http://sourceforge.net/projects/cg-pipeline/ 3. Trimming and filtering run_assembly_trimClean.pl Min length Min avg. quality Min length Min avg. quality 3A. trimming 3B. filtering http://sourceforge.net/projects/cg-pipeline/ More • Software – – – – Fastx toolkit http://hannonlab.cshl.edu/fastx_toolkit/ EA-utils https://code.google.com/p/ea-utils/ AMOS amos: SourceForge.net … and more is out there! • Evaluation – Fabbro et al 2013, “An extensive evaluation of read trimming effects on Illumina NGS data analysis” Algorithms + metrics ASSEMBLY Whole genome sequencing: WGS Large pieces and de novo assembly “Business dog” http://www.buzzfeed.com/tiad/business-dog 52 Whole genome sequencing: WGS Small pieces and reference assembly “Business cat” http://www.quickmeme.com/Business-Cat/ 53 Assembly • Overlaps between reads • Generate contigs (contiguous sequences) • Generate scaffolds NNN N Derive consensus sequence TAGATTACACAGATTACTGA-TTGATGGCGTAA-CTA TAGATTACACAGATTACTGACTTGATGGCGTAAACTA TAG-TTACACAGATTATTGACTTCATGGCGTAA-CTA TAGATTACACAGATTACTGACTTGATGGCGTAA-CTA TAGATTACACAGATTACTGACTTGATGGGGTAA-CTA TAGATTACACAGATTACTGACTTGATGGCGTAA-CTA Derive each consensus base by weighted voting Slide adapted from Andrey Kislyuk, http://www.compgenomics2009.biology.gatech.edu/images/1/12/2009-01-14-compgenomicskislyuk.pdf Recap of assembly reads Paired end reads contigs Scaffold NNNNNNNNNN CG-Pipeline way for Illumina run_assembly reads.fastq.gz –o assembly.fasta • No module yet in CGP for PacBio unfortunately… • Be on the look out for several papers that compare Illumina assemblers. PacBio Assembly • The following slides are courtesy of PacBio Finishing Genomes Using Only PacBio® Reads Hierarchical Genome Assembly Process (HGAP) • Utilizes all PacBio data from single, long-insert library – Longest reads for continuity – All reads for high consensus accuracy • Now available through SMRT® Portal in SMRT Analysis v2.0.1 Chin et al (2013), “Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data” Nature Methods. doi 10.1038/nmeth.2474 Hierarchical Genome Assembly Process (HGAP) 1. Start with long ‘seed’ reads 3. Build consensus 2. Align other reads 4. Construct accurate (>99%) pre-assembled reads HGAP Example - Meiothermus ruber 10 kb SMRTbell™ library 3 SMRT® Cells (C2-C2 Chemistry, PacBio® RS) 250 Mb Long seed reads (>5kb) pre-assembly Pre-assembled long reads Celera Assembler 5 contigs Polish, Quiver 1 contig Collaboration with A. Clum, A. Copeland (Joint Genome Institute) >5 kb HGAP Example - Meiothermus ruber 10 kb SMRTbell™ library 3 SMRT® Cells (C2-C2 Chemistry, PacBio® RS) Long seed reads (>5 kb) pre-assembly Pre-assembled long reads Celera Assembler 5 contigs Polish, Quiver 1 contig Collaboration with A. Clum, A. Copeland (Joint Genome Institute) HGAP Example - Meiothermus ruber 10kb SMRTbell™ library 3 SMRT® Cells (C2-C2 Chemistry, PacBio® RS) Long seed reads (>5 kb) Pre-assembly Pre-assembled long reads Celera Assembler 5 contigs Polish, Quiver 1 contig Collaboration with A. Clum, A. Copeland (Joint Genome Institute) HGAP Example - Meiothermus ruber 10kb SMRTbell™ library 3 SMRT® Cells (C2-C2 Chemistry, PacBio® RS) Long seed reads (>5 kb) Pre-assembly Pre-assembled long reads Celera® Assembler Minimus2 5 contigs Quiver 1 contig 1 contig Collaboration with A. Clum, A. Copeland (Joint Genome Institute) • Single-contig assembly • 99.99965% concordance with reference • 99.3% genes predicted Polish with Quiver for High Accuracy Organism Assembly size (bases) Differences with Sanger reference Meiothermus ruber 3,098,781 11 M. ruber Sanger reference PacBio® reads Targeted Sanger validation SNPs Concordance Nominal validated as Remaining with Sanger QV correct differences reference PacBio calls QV 99.99965% 60 54.5 8 1(3) Estimated Coverage Targets for Finishing Smaller Genomes Assembly Approach / Software Tool Recommended PacBio® Coverage Additional Data Sets Genome Size Constraints None < 10 MB (SMRT Portal) < 130 MB (Command Hierarchical SMRT® Analysis implementation of HGAP (uses Celera® Assembler 75-100X PacBio CLR 7.0) Celera® Assembler via PacBiotoCA (recent compilation) see Koren et al (2013) Line) 75-100X PacBio CLR None 20-50X PacBio CLR 50X short reads ALLPATHS-LG 50X PacBio 3 kb CLR - 50X Illumina® PE - 50X Illumina® jumping libraries MIRA (with PacBiotoCA) 20-50X PacBio CLR 50X short reads 10X PacBio CLR High-confidence contigs Similar to above http://arxiv.org/abs/1304.3752 Hybrid Celera® Assembler 7.0 with PacBiotoCA (SMRT® Analysis) 20 MB Scaffolding AHA (SMRT Analysis) <200 MB; <20,000 contigs Selected publications with PacBio • Application – Katz et al 2013 Mbio “Evolutionary Dynamics of Vibrio cholerae O1 following a Single-Source Introduction to Haiti” – Chin et al 2011 NEJM “The Origin of the Haitian Cholera Outbreak Strain” – Rasko et al 2011 NEJM “Origins of the E. coli Strain Causing an Outbreak of Hemolytic–Uremic Syndrome in Germany” • Assemblers – Chin et al 2013 Nature “Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data” – Koren et al 2012 Nature Biotechnology “Hybrid error correction and de novo assembly of single-molecule sequencing reads” – Bashir et al 2012 Nature Biotechnology “A hybrid approach for the automated finishing of bacterial genomes” The epigenome • PacBio can also detect epigenetic modifications, especially the methylome • Roles for DNA methylation and methyltransferases are uncharacterized in most bacteria • Not your primary task, but it could be a very interesting and novel result of the 2014 compgenomics class Davis et al 2013, Current opinion in microbiology “Entering the era of bacterial epigenomics with single molecule real time DNA sequencing” ASSEMBLY DIFFICULTIES One problem: randomly low coverage (Lander-Waterman) • Assuming random distribution of reads and ignoring repeat resolution issues, • G= genome length • L = length of a single read • N= number of reads sequenced • T= minimum overlap to align the reads together • Then overall coverage is C = LN/G • Coverage for any given base obeys the Poisson distribution: • The number of gaps(bases with 0 coverage) is: A good Lander-Waterman reference: http://www.math.ucsd.edu/~gptesler/186/shotgun_08-handout.pdf Lander and Waterman (1988). Genomic Mapping by Fingerprinting random clones: a mathematical analysis. Genomics 2 pp. 231-239 http://www.cmb.usc.edu/papers/msw_papers/msw-081.pdf Major Problem: repeat elements ? ? Major Problem: repeat elements Major Problem: repeat elements D (1) A (1) B (2) C (2) E (2) G (1) F (1) Unipath graph of the 1.8-Mb genome of C. jejuni Possible paths: ABCDBCEFCEG ABCEFCDBCEG Butler J et al. (1998) ALLPATHS: De novo assembly of whole-genome shotgun microreads. Genome Res. 18:810-820 Comparisons of assemblers • Second-generation – Zhang et al 2011 “A practical comparison of de novo genome assembly software tools for next-generation sequencing technolgies” – Genome Assembly Gold-standard Evaluations (GAGE) http://gage.cbcb.umd.edu/ – Lin et al 2011, “Comparative studies of de novo assembly tools for next-generation sequencing technologies” – Don’t ignore many newer assemblers including SPAdes • Third-generation – Too new! Zhang et al 2011 A quick note on reference assembly • It is possible to map reads to a reference genome using short-read mappers, and then deriving a consensus sequence • Some favorites – Smalt – BWA – Bowtie • Must know how to use samtools for this route Reference assembly notes • To my knowledge no paper exists that compares reference assemblers (could be wrong) • Assembly could be biased – Miss genomic islands – Ends of contigs might be tapered • Good practice for reference assembly: perform de novo assembly on unused reads just in case you missed something Assembly Metrics How do you tell if your assembly is good? Metric description Assembly Length The size of the concatenated assembly Number of Contigs The count of contigs N50 The size of the contig at where half the genome is located in size >N50 and half is located in size <N50 reference Longest Contig Average contig length Kmer21 Frequency of kmers with k=21 GC-content Percentage of the genome that is either G or C Assembly score Log(N50/numberOfContigs) http://www.homolog.us/blogs/blog/2012/06/26/ what-is-wrong-with-n50-how-can-we-make-itbetter-part-ii/ CG-Pipeline/Lee Katz Assembly evaluation QUAST • http://bioinf.spbau.ru/quast The CG-Pipeline way $ run_assembly_metrics.pl assembly.fasta|column –t File assembly.fasta genomeLength 2992976 N50 2992976 numContigs 1 assemblyScore 14.9117787680924 AMOS • amosvalidate • http://amos.sourceforge.net – (build this from source to avoid bugs) Post-assembly manual methods • View the pileups and see if you agree with base calls – – – – Hawkeye (AMOS) IGV viewer Tablet viewer Samtools tview (command line interface) • Compare to other genomes; sort contigs – MAUVE and MAUVE contig mover – Mummer (mummerplot) – Abacas Post-assembly manual methods • Close gaps: Scaffold with Illumina – – – – – – • Scaffold with PacBio – AHA (assembler) GRASS SSPACE SOPRA Babmus2 (AMOS) IMAGE Many others NNN N Acknowledgements • Every single compgenomics class • For help with slides: Eleni Paxinos, Andrew Huang, Amber Schmitke, Maryann Turnsek • For letting me off work, my supervisor Cheryl Tarr • Many, many others who I work with on a daily basis Questions? Lee Katz lkatz@cdc.gov The findings and conclusions in this presentation have not been formally disseminated by the Centers for Disease Control and Prevention and should not be construed to represent any agency determination or policy. The findings and conclusions in this [report/presentation] are those of the author(s) and do not necessarily represent the official position of CDC 86