DCM ERFs ERPs - Wellcome Trust Centre for Neuroimaging
advertisement

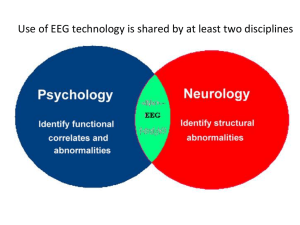
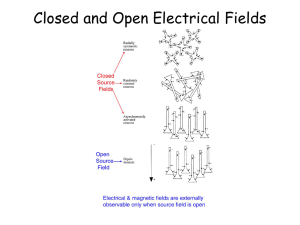
DCM for ERP/ERF: Theory and Practice Methods for Dummies 2013 Helen Pikkat Andrea Gajardo Vidal Overview DCM:Theory • Introduction • DCM • Neural mass model • Bayesian Model Comparison DCM :Practice • SPM analysis • Pre processing Introduction Event related Fields (ERFs) and potentials (ERPs) have been used for decades as putative magnetoand electrophysiological correlates of perceptual and cognitive operations. However, the exact neurobiological mechanism underlying their generation are largely unknown. David et al., (2005), NeuroImage Introduction ERPs • ERPs are measured with electroencephalography (EEG). ERFs • The magnetoencephalography (MEG) equivalent of ERP is the ERF, or event-related field. Dynamic Causal Modelling (DCM) Dynamic Causal Modelling (DCM) is based on an idea initially developed for fMRI data: The measured data are explained by a network model consisting of a few sources, which are interacting dynamically. One can make inferences about connections between sources, or the modulation of connections by task. Ashburner et al. (2013) SPM 8 Manual Dynamic Causal Modelling (DCM) Five essential aspects.. 1. DCMs are dynamic, using (linear or nonlinear) differential equations for describing neuronal dynamics. 2. Second, they are causal in the sense of control theory. 3. Third, DCMs strive for neurophysiological interpretability. 4. Fourth, they use a biophysically motivated and parameterized forward model to link the modelled neuronal dynamics to specific features of measured data. 5. Fifth, DCMs are Bayesian in all aspects. Each parameter is constrained by a prior distribution. Stephan et. al.,(2010), NeuroImage DCM for ERPs and ERFs • The dynamic causal models (DCMs) can explain event-related potentials (ERPs) and fields (ERFs) measured with electroencephalography (EEG) and magnetoencephalography (MEG). DCM for ERPs and ERFs: Types of connectivity Presence of axonal connections Dependence between regions Causal influences between neuronal populations DCM for ERPs and ERFs • DCM for ERPs and ERFs uses the concept of effective connectivity, as opposed to functional connectivity. Effective connectivity refers explicitly to the influence one neuronal system exerts over another. • Effective connectivity is estimated by perturbing the system and measuring the response using Bayesian model inversion. DCM for ERPs and ERFs: The Paradigm Mismatch negativity (MMN): Auditory mismatch negativity Deviant conditions (D) can be embedded in a stream of repeated or standards condition (S) produce a distinct response that can be recorded non-invasively with EEG and MEG. The MMN is the negative component of the waveform obtained by subtracting the event-related response to a standard (common) from the deviant condition (infrequent condition). DCM for M/EEG: Strengths and Limitations STRENGTHS 1.For M/EEG data, DCM is a powerful technique for inferring about parameters that one doesn't observe with M/EEG directly. 2.In general DCM for M/EEG can test hypotheses about what is happening between sources, in a network. 3.One of the most important thing is that DCM combines the spatial forward model with a biologically informed temporal forward model, describing e.g. the connectivity between sources. This critical ingredient not only makes the source reconstruction more robust by implicitly constraining the spatial parameters, but also allows one to infer about connectivity DCM for M/EEG: strengths and limitations LIMITATIONS 1.Model selection: the choosing between two or more alternative models can generate problems. The most common problem is known as “overfitting” (a model fit alone is not a sufficient criterion, as a good fit can often be achieved by simply including a large number of unnecessary parameters). 2. A model that explains only a very small portion of the variance of the data does not inspire much confidence. 3.DCM is capable of identifying the “true” model provided it is among the candidate set? Hierarchical MEG/EEG Neural Mass Model • The majority of neural mass models of MEG/EEG dynamics have been designed to generate spontaneous rhythms (David and Friston, 2003) and epileptic activity (Wendling et al.,2002). These models use a small number of state variables to represent the expected state of large neuronal populations, i.e.the neural mass. • David et al. (2005) developed a hierarchical cortical model to study the genesis of ERFs/ERPs. Hierarchical MEG/EEG Neural Mass Model DCMs for MEG/EEG use neural mass models to explain source activity in terms of the ensemble dynamics of the interacting inhibitory and excitatory subpopulations of neurons, based on the model of Jansen and Rit (1995). This model emulates the activity of a source using three neural subpopulations, each assigned to one of three cortical layers: • • • An excitatory subpopulation in the granular layer. An inhibitory subpopulation in the supragranular layer. An population of deep pyramidal cells in the infra-granular layer. Hierarchical MEG/EEG Neural Mass Model The excitatory pyramidal cells receive excitatory and inhibitory input from local interneurons (via intrinsic connections), and send excitatory outputs to remote cortical areas via extrinsic connections. Using these connection rules, it is straightforward to construct any hierarchical cortico-cortical network model of cortical sources. Hierarchical MEG/EEG Neural Mass Model Bottom-up or forward connections • that originate in agranular layers and terminate in layer 4. Top-down or backward connections • that connect agranular layers. Lateral connections • that originate in agranular layers and target all layers. + Canonical MicroCircuit (CMC) Hierarchical MEG/EEG Neural Mass Model • The cortex has a hierarchical organization (Crick and Koch, 1998; Felleman and Van Essen, 1991). • Forward, backward and lateral processes that can be understood from an anatomical and cognitive perspective (Engel et al., 2001). • DCM for ERPs and ERFs takes the spatial forward model into account. This makes DCM spatiotemporal model of the full data set (over channels and peri-stimulus time). Hierarchical MEG/EEG Neural Mass Model Bayesian Model Comparison . The inversion of a DCM provides information about the underlying cortical pathways and their causal architecture. SPM presentation (2012) Bayesian Model Comparison • What model fit better your data? • Bayesian model comparison (Penny et al., 2004) selects the model, among competing models, that best explains the data. • Given equal prior probabilities for the models considered, they are compared using their marginal likelihood or evidence for each model. Bayesian Model Comparison A B • How to compare models to data? • What makes model A better than model B? – If it describes the data better… – What do we mean by “describing better”? Bayesian Model comparison Model comparison for group studies Random effect (RFX) Fixed effect (FFX) For multi-subject analyses, two options exist depending on whether one assumes that the parameters of interest are fixed effects in the population (FFX) or are themselves probabilistically distributed in the population (RFX). Bayesian Model comparison Fixed effect (FFX) In the FFX case, one assumes that the optimal model is the same for each subject in the population. This assumption is warranted when studying a basic physiological mechanism that is unlikely to vary across the subjects sampled. Bayesian Model comparison Random effect (RFX) On the other hand RFX accounts for heterogeneity of model structure across subjects and yields posterior model probabilities and exceedance probabilities. In either case, model space partitioning and subsequent comparison of model families (family-level inference) should be considered when the hypothesis of interest concerns model structure and not any particular model parameter. DCM for ERP/ERF: practice There are different DCMs for EEG/MEG data This presentation focuses on ERPs (based in large part on the sample SPM dataset for MMN) DCM for ERP/ERFs recquires an experimental manipulation. Steps to follow Hypothesis pre-DCM Preprocessing Model specification Model inversion Model comparison DCM pre-DCM Hypothesis Preprocessing • What kind of hypothesis can you test? • * about the model space (e.g. importance of specific areas) • * about the model parameters (e.g. connection strength, direction) • Preprocessing is done as for any other EEG data analysis... DCM: SPM DCM: SPM - data & design DCM: SPM - data & design Choose the period you want to model Choose the neural model (ERP, CMC, ...) Choose the type of DCM Insert the conditions Choose the number of components Specify contrasts for your conditions Model specification What cortical areas and connections might explain the data? Model specification * Specify the areas for each model Based on: - the literature - fMRI data - the previous or current EEG data Note that you can define models with different number of regions (which is not possible in DCM for fMRI). * Specify the connections for each model Based on: - anatomical/physiological knowledge Model specification + a matrix for backwards connections + a matrix for lateral connections + a matrix for connections that are allowed to vary between conditions (modulatory effects) Model specification: SPM Model specification: SPM Name the sources Insert the coordinates of the sources (in MNI coordinates) Specify the connections Model inversion Finds the parameters that minimize differences between observed measurements and the predicted models. As a result you will get: - posterior distribution of parameters You can plot the data for ERPs and model fit for each mode. Model inversion Predicted data (model) Data Expectation-Maximization algorithm Compute model response using current set of parameters Compare model response with data Improve model parameters if possible (until the maximum likelyhood is found) Result: posterior distributions of parameters Make inferences on parameters Model inversion: SPM Then specify the connections for the other model(s) and estimate the parameters for these Check the results from here Estimate the model Model comparison Remember - there is no right model! We can get the information for: 1. The best model (BMS) 2. The coupling parameters of this model Usually, BMS (Bayesian Model Selection) is first used to select an optimal model from all alternatives, and then the posterior or conditional inferences about the parameters of this optimal model are reported. Model comparison: BMS BMS (Bayesian Model Selection) computes the model evidence (the probability of data) given some model and priors. - The most likely model is the one with the largest log-evidence. - Conventionally: log-evidence greater then 3 provides strong evidence. *If you can't differentiate two models then they can be either too similar or the data might be too noisy ot they might not have fitted well. * If you were interested only in model structure you can just use BMS and don't have to interpret the results from mode estimation Model comparison: BMS in SPM Compare models Model comparison: BMS in SPM Model comparison: BMS in SPM Results: - a bar plot of the log-model evidences for all models Model comparison: BMS in SPM Results: - the probability, for each model, that it produced the data. Back to model inference When you are interested in specific parameters in the model(s) use the results from the model estimation. Use BMA (Bayesian model averaging) to compare parameters for different models. Final notes • - DCM can tell you about the changes in connectivity between sources. • - ...but it can only compare the models that you provide • - Therefore your models should be based on your hypothesis and be plausible • - Any result obtained is relative and dependent on the models that you defined • Thank very much to our expert Harriet Brown!! Reference List den Ouden, D-B., Saur, D., Mader, W., et al., (2012), Network modulation during complex syntactic processing, Neuroimage, 59-1, p.815-823 David, O., Kiebel, S.J., Harrison, L.M., et al., (2006), Dynamic causal modeling of evoked responses in EEG and MEG, Neuroimage, 30-4, p.1255-1272 Kiebel, S. J., Garrido, M. I.,; Moran, R. J., et al., (2008), Dynamic causal modelling for EEG and MEG, Cognitive Neurodynamics, 2-2, p.121-136 David, O., Harrison, L., Friston, K.J., (2005), Modelling event-related responses in the brain, Neuroimage, 25-3, p.756-770 Lohmann, G., Erfurth, K., Mueller, K. et al., (2012), Critical comments on dynamic causal modelling, Neuroimage, 59-3, p.2322-2329 SPM8 Manual (2013) Litvak, V. (2012, May). The principles of DCM. Talk given at SPM course London. Daunizeau, J. (2012, May). DCM for evoked responses. Talk given at SPM course London. Bastos, A. & Dietz, M., (2012, May). Demo - DCM. Talk given at SPM course London. Stephan, K. E., Penny, W. D., Moran, R. J., Den Ouden, H. E. M., Daunizeau, J., & Friston, K. J. (2010). Ten simple rules for dynamic causal modeling. NeuroImage, 49(4), Previous MfD talks Reference List • • • • • • • David, O., Harrison, L., & Friston, K. ( 2005). Modelling event-related responses in the brain. Neuroimage, 25, 3, 756-770. David, O., Kiebel, S. J., Harrison, L. M., Mattout, J., Kilner, J. M., & Friston, K. J. (2006). Dynamic causal modeling of evoked responses in EEG and MEG. Neuroimage, 30, 4, 1255-1272 Friston, K. J., Harrison, L., & Penny, W. (2003). Dynamic causal modelling. Neuroimage, 19, 4, 1273-1302. Garrido, M. I., Kilner, J. M., Kiebel, S. J., Stephan, K. E., & Friston, K. J. (2007). Dynamic causal modelling of evoked potentials: a reproducibility study. Neuroimage, 36, 3, 571-80 Stephan, K. E., Penny, W. D., Moran, R. J., den, O. H. E. M., Daunizeau, J., & Friston, K. J. (2010). Ten simple rules for dynamic causal modeling. Neuroimage, 49, 4, 3099-3109. Ashburner et. al., (2012). SPM 8 MANUAL. Wellcome Trust Centre For Neuroimaging. http://www.fil.ion.ucl.ac.uk/spm/course/