the math
advertisement

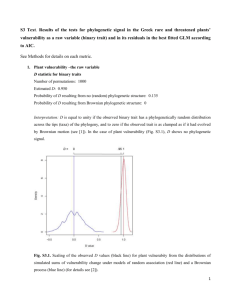
Evolutionary processes that can lead to change or stasis over time Pattern: Change Stasis Adaptive Directional selection • Stabilizing selection • Fluctuating selection (noise with no trend) Nonadaptive Mutation Genetic drift • Lack of genetic variation • Constraint (?) • Antagonistic correlations among traits under selection • Swamping by gene flow Blomberg’s K – measure of phylogenetic signal phylogenetic signal low K = 0.18 Data diagnostics brownian K~1 high K = 1.62 Blomberg et al. 2003 Evolution examples from Ackerly 2009 PNAS K>1 Brownian motion – assumptions and interpretations Evolutionary models Brownian motion – assumptions and interpretations ∞ -∞ Evolutionary models Ornstein-Uhlenbeck model (OU-1) the math: brownian motion + ‘rubber band effect’ change is unbounded (in theory), but as rubber band gets stronger, bounds are established in practice 5 0 -5 -15 trait value 10 15 repeated movement back towards center erases phylogenetic signal, leading to K << 1 0 50 100 150 200 250 300 time Evolutionary models see Hansen 1997 Evolution Butler and King 2004 Amer. Naturalist Ornstein-Uhlenbeck model (OU-1) the math: brownian motion + ‘rubber band effect’ change is unbounded (in theory), but as rubber band gets stronger, bounds are established in practice 5 0 -5 -15 trait value 10 15 repeated movement back towards center erases phylogenetic signal, leading to K << 1 0 50 100 150 200 250 300 time Evolutionary models see Hansen 1997 Evolution Butler and King 2004 Amer. Naturalist Ornstein-Uhlenbeck model (OU-2+) the math: brownian motion + ‘rubber band effect’ with different optimal trait values for clades in different selective regimes Balance of stabilizing selection within clades vs. how different the optima are can lead to strong or weak phylogenetic signal This example would be VERY strong signal Evolutionary models see Hansen 1997 Evolution Butler and King 2004 Amer. Naturalist Early-burst model the math: brownian motion with a declining rate parameter change is unbounded (in theory), but divergence happens rapidly at first and then rates decline and lineages change little divergence among major clades creates high signal: K >> 1 Evolutionary models Harmon et al. 2010 Assign proportional weighting of alternative models that best fit data Harmon et al. 2010 Rates of phenotypic diversification under Brownian motion 1 felsen = 1 Var(loge(trait)) million yrs time var(x) Rates of phenotypic diversification under Brownian motion higher rate lower rate time var(x) Diversification of height in maples, Ceanothus and silverswords rate = 0.015 felsens 0.10 felsens 0.83 felsens ~5.2 Ma ~30 Ma ~45 Ma Evolutionary rates Ackerly 2009 PNAS Rates of phenotypic diversification (estimated for Brownian motion model) Rate (felsens) North temperate California Hawai’i Height Leaf size ±1 s.e. Ackerly, PNAS in review time var(x) Linear parsimony 0 Squared change parsimony = ML with BL =1 2.44 0.08 2 1.32 1 0.52 0 0.24 0 0.12 0 ML with BL as shown 0.096 2.54 1.6 0.96 0.67 0.56 ML with BL as shown C F Node ML estimate Lower 95% CI Upper 95% CI A 0.56 -0.77 1.89 B 0.67 -0.43 1.78 C 0.096 -0.61 0.81 D 0.96 0 1.95 E 1.6 0.76 2.45 F 2.54 1.86 3.2 E D B A Oakley and Cunningham 2000 Polly 2001 Am Nat Independent contrasts a 2 8 6 16 1 6 b R = 0.74 11 14 8.5 9 11.5 11 19 18 13 12 a 2 8 6 16 1 6 11 14 8.5 9 11.5 11 10 8 4 8 4 12 19 18 13 12 3 2 6 10 -6 -6 10 10 16 15 2 -2 6 5 5 11 13 17 8 6 9 14 b R = 0.74 c R = 0.92 Oakley and Cunningham 2000 Oakley and Cunningham 2000 A21223 Fig. 2 1) Assume bivariate normal distribution of variables with = 0 Distribution of correlation coefficients (R) under null hypothesis 2) Draw samples of 22 and calculate correlation coefficient Crit(R, = 0.05, df = 20) is 0.423 3) Repeat 100,000 times! N < -0.423 = 2519; N > 0.423 = 2551 Type I error = 0.051 1) Assume bivariate normal distribution of variables with = 0.5 Crit(R, = 0.05, df = 20) is 0.423 2) Draw samples of 22 and calculate correlation coefficient N < -0.423 = 5 N > 0.423 = 68858 3) Repeat 100,000 times! Power = 0.69 1) Assume bivariate normal brownian motion evolution along a phylogeny, with ~ 0.0 2) Calculate R using normal correlation coefficient 3) Repeat 10,000 times! Crit(R, = 0.05, df = 20) is 0.423 N < -0.423 = 1050 N > 0.423 = 1044 Type I error = 0.21 1) Assume bivariate normal brownian motion evolution along a phylogeny, with ~ 0.0 2) Calculate R using independent contrasts 3) Repeat 10,000 times! Crit(R, = 0.05, df = 20) is 0.423 N < -0.423 = 246 N > 0.423 = 236 Type I error = 0.048 From Ackerly, 2000, Evolution