E. coli
advertisement
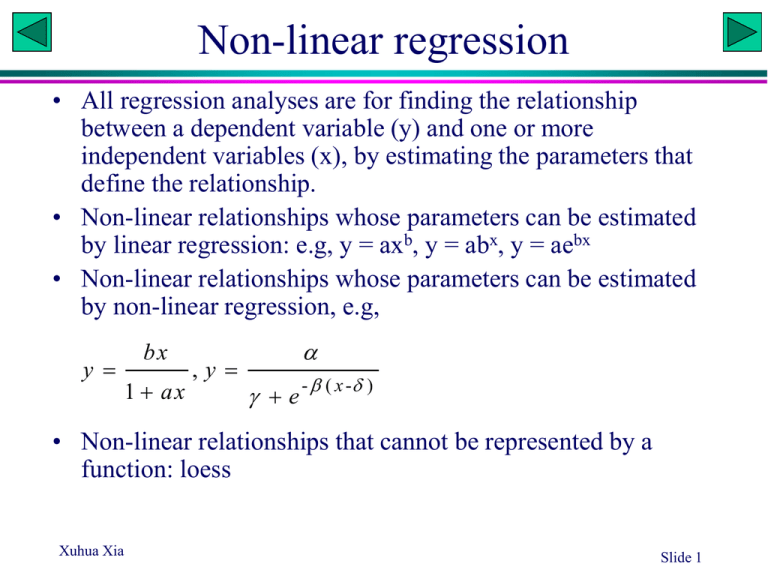
Non-linear regression • All regression analyses are for finding the relationship between a dependent variable (y) and one or more independent variables (x), by estimating the parameters that define the relationship. • Non-linear relationships whose parameters can be estimated by linear regression: e.g, y = axb, y = abx, y = aebx • Non-linear relationships whose parameters can be estimated by non-linear regression, e.g, y bx 1 ax ,y e - ( x - ) • Non-linear relationships that cannot be represented by a function: loess Xuhua Xia Slide 1 Growth curve of E. coli • A researcher wishes to estimate the growth curve of E. coli. He put a very small number of E. coli cells into a large flask with rich growth medium, and take samples every half an hour to estimate the density (n/L). • 14 data points over 7 hours were obtained. • What is the instantaneous rate of growth (r). What is the initial density (N0)? • As the flask is very large, he assumed that the growth should be exponential, i.e., y = a·ebx (Which parameter correspond to r and which to N0?) • Three approaches – Log-Transform to linear relationship – Direct least-square solution (EXCEL solver) – Direct least-absolute-difference solution (EXCEL solver) Xuhua Xia Time Density 1 20.023 2 39.833 3 80.571 4 161.102 5 317.923 6 635.672 7 1284.54 8 2569.43 9 5082.65 10 10220.8 11 20673.9 12 40591.4 13 81374.6 14 163964 Slide 3 Scatter plot 180000 D D0e Density 160000 y = 10.016e0.6928x R2 = 1 rt 140000 In EXCEL: 120000 Log-transform D Run linear regression Obtain D0 and r 100000 80000 60000 40000 20000 0 1 3 5 7 9 11 13 Time Xuhua Xia Slide 4 EXCEL solver Time Density Pred SS a 9.554915 Pred SAD a 9.554956 1 20.023 39.833 80.571 161.102 317.923 635.672 1284.54 2569.43 5082.65 10220.8 20673.9 40591.4 81374.6 163964 19.172 0.724 b 0.696402 19.173 0.850 b 0.696453 38.469 1.860 38.473 1.360 77.189 11.436 77.201 3.370 154.882 38.690 154.914 6.188 310.774 51.115 310.854 7.069 623.573 146.380 623.767 11.905 1251.212 1111.019 1251.666 32.878 2510.582 3463.120 2511.621 57.809 5037.532 2036.008 5039.875 42.779 10107.907 12739.579 10113.126 107.651 20281.716 153787.323 20293.226 380.647 40695.664 10862.758 40720.843 129.404 81656.653 79530.444 81711.360 336.718 163845.689 13967.378 277747.832 163963.851 0.022 1118.648 2 3 4 5 6 7 8 9 10 11 12 13 14 Get initial value for r: D2 D1 D0e rt 2 D0e rt1 D0e r ( t1 1) D0e rt1 rt D0e 1 e D0e rt1 r e r Initial value for D0 is obtained with t = 0 Xuhua Xia Slide 5 Body weight of wild elephant • A researcher wishes to estimate the body weight of wild elephants. • He measured the body weight of 13 captured elephants of different sizes as well as a number of predictor variables, such as leg length, trunk length, etc. Through stepwise regression, he found that the inter-leg distance (shown in figiure) is the best predictor of body weight. • He learned from his former biology professor that the allometric law governing the body weight (W) and the length of a body part (L) states that W = aLb • Use the three approaches to fit the equation Xuhua Xia Slide 6 Scatter plot W = aLb In EXCEL: 50 Log-transform W and L Run linear regression Obtain a and b 40 W y = 20.018x 2.1382 R2 = 0.9955 30 20 10 0 0.2 0.4 0.6 0.8 1 1.2 1.4 L Xuhua Xia Slide 7 EXCEL solver L 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 1.1 1.2 1.3 1.4 1.5 Xuhua Xia W 1.657 2.500 4.680 7.075 10.070 11.988 14.836 18.318 23.496 27.897 36.796 44.611 50.183 Pred 1.165 2.285 3.853 5.904 8.471 11.580 15.258 19.527 24.409 29.924 36.093 42.932 50.459 251.859 SS 0.242 0.046 0.684 1.370 2.557 0.166 0.178 1.461 0.833 4.111 0.495 2.820 0.076 15.039 a 19.52661 b 2.341457 W=aLb Initial values: W2 W1 L 2 aL L1 b 2 b 1 b aL W L ln 2 b ln 2 W1 L1 a W b L Slide 8 DNA and protein gel electrophoresis • How to estimate the molecular mass of a protein? – A ladder: proteins with known molecular mass – Deriving a calibration curve relating molecular mass (M) to migration distance (D): D = F(M) – Measure D and obtain M • The calibration curve is obtained by fitting a regression model Xuhua Xia Slide 9 Protein molecular mass • The equation D=aebM appears to describe the relationship between D and M quite well. This relationship is better than some published relationships, e.g., D = a – b ln(M) • The data are my measurement of D and M for a subset of secreted proteins from the gastric pathogen Helicobacter pylori (Bumann et al., 2002). • Homework: use the data and the three approaches to estimate parameters a and b (You don’t need to submit) Mass D 5 14.5 10 12.6 20 9.4 30 7.1 40 5.3 50 3.9 60 3.05 70 2.3 80 1.75 Bumann, D., Aksu, S., Wendland, M., Janek, K., Zimny-Arndt, U., Sabarth, N., Meyer, T.F., and Jungblut, P.R., 2002, Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 70: 3396-3403. Xuhua Xia Slide 10 Area and Radius What is the functional relationship between the area and the radius? Homework (you do not need to submit): Measure the area A (by counting the squares) and radius r for each circle and estimate the parameters c and d in the equation A = crd by using the three approaches. Xuhua Xia Toxicity study: pesticide 100 90 Percentage killed 80 70 60 50 40 30 20 10 0 25 30 35 40 45 50 55 60 65 70 Dosage What transformation to use? Xuhua Xia Slide 12 Probit and probit transformation • Probit has two names/definitions, both associated with standard normal distribution: – the inverse cumulative distribution function (CDF) – quantile function 0.9 0.8 0.7 0.6 CDF • CDF is denoted by (z), which is a continuous, monotone increasing sigmoid function in the range of (0,1), e.g., (z) = p (-1.96) = 0.025 = 1 - (1.96) • The probit function gives the 'inverse' computation, formally denoted -1(p), i.e., probit(p) = -1(p) probit(0.025) = -1.96 = -probit(0.975) • [probit(p)] = p, and probit[(z)] = z. 1 0.5 0.4 0.3 0.2 0.1 0 -2.5 -1.5 -0.5 0.5 1.5 z Xuhua Xia Slide 13 Data Dosage Xuhua Xia 27 28 31 31 35 36 37 38 38 40 41 43 44 44 44 45 45 45 46 46 46 47 47 48 49 49 49 49 50 50 51 %Killed 0.9 1.39 2.4 2.49 6.42 7.78 9.16 10.21 11.71 16.24 16.9 22.94 27.35 27.45 28.14 28.97 29.96 30.5 34.3 35.39 35.65 37.55 38.46 40.97 44.37 45.71 46.66 47.38 49.86 52.26 55.12 Probit Pred PredOriginalSUMMARY OUTPUT -2.365618 -2.354331 0.927805 -2.200097 -2.251524 1.217619 Regression Statistics -1.977368 -1.943104 2.600181 Multiple R 0.999559 -1.961678 -1.943104 2.600181 R Square 0.999118 -1.520442 -1.531877 6.277642 Adjusted R Square 0.999103 -1.420026 -1.42907 7.649205 Standard Error 0.029949 -1.330967 -1.326263 9.237624 Observations 60 -1.269676 -1.223457 11.05786 -1.189609 -1.223457 11.05786 ANOVA -0.984642 -1.017843 15.43762 df -0.958124 -0.915036 18.00863 Regression -0.740824 -0.709423 23.9031 Residual -0.602262 -0.606616 27.20528 Total SS MS 1 58.94878 58.94878 F 65722.54954 Significance F 2.71096E-90 58 0.052022 0.000897 59 59.00081 -0.599259 -0.606616 27.20528 -0.578688 -0.606616 27.20528 Coefficients Standard Error t Stat P-value -0.554261 -0.50381 30.71976 Intercept -5.130112 0.020381 -251.7115 7.83524E-90 -0.525551 -0.50381 30.71976 Dosage 0.102807 0.000401 256.3641 2.71096E-90 -0.510073 -0.50381 30.71976 -0.404289 -0.401003 34.4209 -0.374812 -0.401003 34.4209 -0.36783 -0.401003 34.4209 -0.317321 -0.298196 38.27768 -0.293421 -0.298196 38.27768 -0.228317 -0.195389 42.25441 -0.141595 -0.092583 46.31176 -0.107742 -0.092583 46.31176 -0.083819 -0.092583 46.31176 -0.065721 -0.092583 46.31176 -0.003509 0.010224 50.40788 0.05668 0.010224 50.40788 0.128694 0.113031 54.49969 Slide 14 Non-linear regression • In rapidly replicating unicellular eukaryotes such as the yeast, highly expressed intron-containing genes requires more efficient splicing sites than lowly expressed genes. • Natural selection will operate on the mutations at the slicing sites to optimize splicing efficiency. • Designate splicing efficiency as SE and gene expression as GE. • Certain biochemical reasoning suggests that SE and GE will follow the following relationships: Xuhua Xia GE SE 1 0.46 2 0.47 3 0.57 4 0.61 5 0.62 6 0.68 7 0.69 8 0.78 9 0.7 10 0.74 11 0.77 12 0.78 13 0.74 13 0.8 15 0.8 16 0.78 Slide 16 Scatter plot 0.9 0.8 SE 0.7 0.6 SE 0.5 GE 1 GE 0.4 0.3 0 2 4 6 8 10 12 14 16 GE Initial values: Xuhua Xia 0.4 (inferred when GE = 0) / 1 or (inferred when GE is very large) When GE = 8, we have (0.4+8 )/(1+8 ) = 0.78 Slide 17 EXCEL: Solver GE 1 2 3 4 5 6 7 8 9 10 11 12 13 13 15 16 SE 0.46 0.47 0.57 0.61 0.62 0.68 0.69 0.78 0.7 0.74 0.77 0.78 0.74 0.8 0.8 0.78 Pred 0.436655 0.510256 0.565294 0.608005 0.642114 0.669981 0.693177 0.712784 0.729577 0.74412 0.756837 0.768052 0.778016 0.778016 0.794944 0.802195 Xuhua Xia SS Alpha 0.333196 0.000544981 Beta 0.192031 0.00162052 Gamma 0.202841 2.21506E-05 3.98053E-06 0.000489015 0.000100378 1.00918E-05 0.004517926 0.000874801 1.69749E-05 0.000173259 0.000142753 0.001445212 0.000483299 2.55629E-05 0.000492612 0.010963515 SE GE 1 GE Slide 18