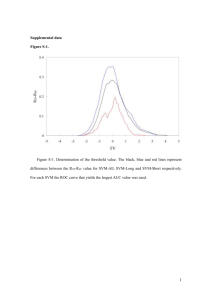
2. Feature Selection
advertisement

Strathclyde Hyperspectral Imaging Centre About the centre Academic: Professor and lecturer Research: 2 post docs (sponsored by Argans and TiC) 2 PhD students (sponsored by Gilden and AZ) working on Fundamental research and HSI applications HSI imaging facility donated by Gilden Photonics HSI Community building activities & partners 1st & 2nd UK Hyperspectral Imaging Conferences Strathclyde April 2010 May 2011 Sponsors/ Exhibitors Gilden Photonics, Gooch & Housego, Specim, Pacer, PANDA, Headwall, Photometrics, Hamamatsu, Mapping Solutions Academic Partners, D Pullan, Univ Leicester, A Harvey, Herriot Watt , D Reynolds UWE, S Morgan, Nottingham 3rd HSI Conference INGV, Rome 15-16 May 2012 HSI Projects Applications Fundamental research Algae classification New data processing tools- Copulas Beef quality assessment Drug dissolution process Finger print detection Chinese tea classification Band selection /data reduction methods Quantitative Assessment of Beef Quality with Hyperspectral Imaging using Machine Learning Techniques Jinchang Ren1, Stephen Marshall 1, Cameron Craigie2,3 and Charlotte Maltin4 1Hyperspectral Imaging Centre, University of Strathclyde, Glasgow, U. K. 2Scottish Agricultural College (SAC), Edinburgh, U. K. 3IVABS, Massey University, Palmerston North, N. Z. 4Quality Meat Scotland, Ingliston Newbridge, U. K. Outline 1. Introduction 2. Feature Selection 3. Machine Learning for Prediction 4. Experiments Results and Discussions 5. Conclusions 1. Introduction NIR Hyperspectral Imaging System [14] Hyperspectral imaging of meat in NIR spectral ranges To predict meat quality of •Freshness •Tenderness •Fat •Protein •Moisture content Image processing and machine learning techniques like neural network are used for prediction & classification. Nearly non-invasive and accurate 1. Introduction –Existing Work NIR sampling over 700-2500nm is preferred to allow spectra acquired in three modes, reflection, transmission and transflection, and also enables response to key food components like C-H, O-H and N-H molecular bonds. 1) Qiao et al [6] used selected bands on a trained neural network for prediction of drip loss, pH and surface colour of pork, and the prediction correlation coefficients are 0.77, 0.56 and 0.86. 2) Eimasry [7] employed PCA for data reduction and partial least square regression for prediction of surface colour, tenderness and pH of beef. The correlations are 0.88, 0.73 and 0.81; 3) Carigie [8] used similar linear regression on beef data captured under commercial conditions and the correlation is less than 35% for surface colour, pH and M. longissimus dorsi tenderness. 2. Feature Selection – Spectral Profiles Usually the spectral profiles are used as features yet some spectral bands or their combinations are more distinguishable than others thus feature selection is desirable. 2. Feature Selection 1) Spectral redundancy makes it possible for feature selection and data reduction; 2) Band selection is not used here as though it is found to yield good results [1, 10]. Instead, principal component analysis (PCA) is utilised as it is a standard one which has been integrated in many existing development tools like Matlab. 3) We apply PCA to the original spectral data and then select the dominant principal components as new features for training and prediction. 3. Machine Learning for Prediction 1) Linear regression and its variations are widely used for prediction of relevant parameters, such as partial least square regression (PSLR) in [7], the general linear models (GLM) in [8] and multi-linear regression (MLR) in [12]. 2) Since the parametric models are not necessary linear, machine learning approaches is preferred where support vector machine (SVM) is employed. 3) SVM enables effective regression and prediction for both linear and non-linear cases and it general outperforms neural network in machine learning. 3. Support Vector Machine (SVM) f SVM ( x ) w ( x ) b T Parameters w and b respectively refer to a weight vector and a bias that can be determined in the training process through minimizing a cost function, and Φ refers to a (nonlinear) mapping to map the input vector x into a higher dimensional space for easily separated by a linear hyperplane as illustrated below. This figure illustrates the concept of SVM where a non-linearly-separable problem becomes linearly separable in the mapped feature space. 3. SVM- Training K f (x ) K (x, sk ) b SVM k 1 T K ( x , s k ) ( x ) ( s k ) 3. SVM- Kernel Functions & Evaluation K (x i , x j ) x i x j linear kernel T K ( x i , x j ) ( x i x j 1) T K (x i , x j ) e xi x j 2 polynomina l kernel p /( 2 ) 2 RBF kernel ˆ ,Y ) C ( Y svm gt LibSVM is used for the implementation of the SVM in both training and prediction. S y yˆ S yy S yˆ yˆ n S yy i 1 S yˆ yˆ n ˆ ( i )] n [ Yˆ ( i )] 2 [ Y svm svm 2 i 1 [Y i 1 1 i 1 n S y yˆ 2 i 1 n The correlation is defined to the right for evaluation. n [Y gt ( i )] n [ Y gt ( i )] 1 2 gt n n i 1 i 1 1 ( i )Yˆsvm ( i )] n [ Y gt ( i )][ Yˆsvm ( i )] 4. Experimental Datasets 1) Our data is the one used in [8] captured under commercial conditions; 2) The data is acquired using a NIR spectrometer at Quality Meat Scotland [15] and the spectrum ranges from 350nm to 1800nm with a spectral resolution of 1nm; 3) The mean and median values in the absorbance form from 10 repetitive scans are recorded for processing. 4) The data samples are 234 in total. 5) A 10-fold cross validation scheme is used for evaluation. 4. Experimental Results Table 1: Correlation of predicted M. longissimus dorsi shear force values to the ground truth under various kernel functions and number of principal components. Correlation Number of principal components = 13 Number of principal components = 20 linear polynomial RBF 0.298 0.450 0.396 0.305 0.533 0.288 4. Analysis of Results 1) Polynomial kernel function produces the best results; and increasing the number of principal components helps to improve the prediction. 2) Linear kernel function and RBF kernel function tend to yield much worse results. Increasing the number of principal components does not certainly improve the results from RBF kernel, though slightly better results can be found for linear kernel. 3) This on one hand indicates that polynomial kernel function seems the best choice for this application. On the other hand, it explains why linear models used in [8] generated the poor results. 4. Experimental Results For some principal components, the correlation values are as high as 0.20.25! It is possible to choose useful principal components for data prediction, and this will be investigated in the next stage. Fig. 3. Correlation values between the ground truth and the extracted principal components. 4. Conclusions 1) We have applied SVM for data prediction in food quality analysis and found polynomial kernel to yield the best results; 2) Using PCA for dimension reduction, we have significantly improved the prediction of M. longissimus dorsi shear force correlation values from less than 0.35 to about 0.533; 3) How to apply our approach to predict other parameters and also to evaluate the possibility in choosing principal components for prediction will be investigated in the near future. Acknowledgements The authors would like to thank colleagues in industry and the Scottish Agricultural College for their help in providing and collecting the data. Thank you for your attention! Any Questions?