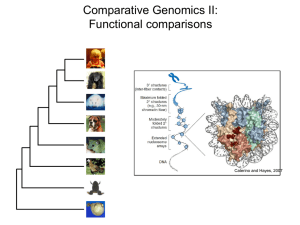
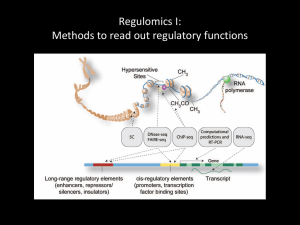
1 Distant-Acting Enhancers
advertisement

Genome-wide Identification of Craniofacial Transcriptional Enhancers Axel Visel Scientist, Genomics Division Lawrence Berkeley National Laboratory Outline 1 Distant-Acting Enhancers Why are they important? How can we find them in the genome (and determine their function)? 2 Finding Developmental Face/Palate Enhancers Data from preliminary ChIP-seq and transgenic mouse studies 3 FaceBase – Contributions and Expectations Data and reagents Interactions Enhancers are Required for Development distant-acting enhancers promoter proteincoding wild-type limb enhancer deleted limb enhancer Sagai et al. 2005 1 megabase Shh gene mouse embryo Lettice et al. 2003 Lettice et al. 2003 human Shh enhancer point mutations Noncoding Sequences in Human Disease Meta-Analysis of Genome-Wide Association Studies (GWAS): Distant Enhancers? 60% Linked to Exons 40% Noncoding LD blocks Of 1,200 disease-associated SNPs, 40% are not linked to any coding gene Visel/Pennacchio/Rubin 2009 (Nature 461:199) How do we find enhancers? Approach A: Extreme Conservation of Non-coding sequences mouse inject into fertilized mouse egg fugu PCR amplify clone P LacZ pHsp68LacZ >500 enhancers identified to date see http://enhancer.lbl.gov reimplant collect at e11.5 LacZ staining Major limitation: can’t find enhancers active in a particular process, e.g. face development minimum reproducibility: 3 embryos How do we find enhancers? Approach B: ChIP-seq with the enhancer-associated p300 protein microdissection midbrain tissue forebrain tissue limb tissue limb p300 ChIP-Seq forebrain midbrain 2,400,000 reads 3,600,000 reads 3,500,000 reads 2,100 peaks 2,400 peaks 600 peaks mouse embryo (e11.5) Test in transgenic mouse assay (Nature 457:854, 2009) p300 ChIP-Seq Predicts in vivo Enhancer Activity ChIP-seq forebrain midbrain limb transgenic mouse assay 80%-90% success rate (n>100) 11/12 5/5 8/8 Enables accurate predictions of genomic locations AND activity of enhancers 11/11 (Nature 457:854, 2009) Outline 1 Distant-Acting Enhancers Why are they important? How can we find them in the genome (and determine their function)? 2 Finding Developmental Face/Palate Enhancers Data from preliminary ChIP-seq and transgenic mouse studies 3 FaceBase – Contributions and Expectations Data and reagents Interactions Enhancers Play a Role in Clefting Disorders enhancer SNP disrupts a single AP-2a binding site x SNP human chr1 IRF6 gene human cleft lip and palate associated with cleft lip/palate virtual section of mouth region enhancer activity in ectoderm of fusing facial prominences Rahimov et al. 2008 (Nature Genetics 40:1341) Jeff Murray Lab, OPT data: David FitzPatrick ChIP-seq for Craniofacial Enhancer Discovery Example: Enhancer near known clefting gene MSX1 mx mx mx Msx1 gene expression in maxillary component of 1st branchial arch (Mackenzie et al., Development 111:269) Three-dimensional imaging of enhancer activity Optical Projection Tomography (Sharpe et al., Science 296:541) OPT of Enhancer Browser embryos: David FitzPatrick/Harris Morrison, MRC Edinburgh Three-dimensional imaging of enhancer activity Optical Projection Tomography (Sharpe et al., Science 296:541) OPT of Enhancer Browser embryos: David FitzPatrick/Harris Morrison, MRC Edinburgh In vivo validation of ChIP-seq predictions OPT scans: David FitzPatrick/Harris Morrison, MRC Edinburgh Outline 1 Distant-Acting Enhancers Why are they important? How can we find them in the genome (and determine their function)? 2 Finding Developmental Face/Palate Enhancers Data from preliminary ChIP-seq and transgenic mouse studies 3 FaceBase – Contributions and Expectations Data and reagents Interactions Visel Lab – FaceBase Aims Genome-wide identification of enhancer candidates p300 ChIP-seq: timepoints (e11.5 – e15.5), better spatial resolution large-scale sequencebased data RNA-seq data Transgenic validation and characterization test 30 candidate sequences/year in transgenic mice whole-mount photos and OPT data (collaboration with FitzPatrick lab) provide validated vectors as reagents to other FaceBase investigators image/video/3D data Follow-up of human genetic studies test risk alleles of clefting-associated craniofacial enhancers in mice integration of enhancer data with human genetic data Visel Lab – FaceBase Expectations Developmental biology and expression imaging groups Intersect with gene expression data Please approach us with regions of interest! Human genetics groups Please approach us with non-coding cleft-associated regions! Use ChIP-seq and transgenics to search for enhancers Transgenic testing of cleft-associated risk variants Acknowledgments Lawrence Berkeley National Lab and DOE Joint Genome Institute Len Pennacchio Eddy Rubin Matt Blow Shyam Prabhakar Mouse Transgenics Malak Shoukry Jennifer Akiyama Veena Afzal Amy Holt Ingrid Plaijzer-Frick Roya Hosseini Collaborators/Contributors: Terri Beaty, Robert Cornell, Michael Dixon, David FitzPatrick, Rulang Jiang, Michael Lovett, Mary Marazita, Jeff Murray, Stephen Murray, Leif Oxburgh, Bing Ren, John Rubenstein, Brian Schutte, Alan Scott, Douglas Spicer http://enhancer.lbl.gov Next-Gen Sequencing NIDCR (FaceBase) Tao Zhang Feng Chen Crystal Wright Enhancer Browser Inna Dubchak Simon Minovitsky DOE, NHGRI