Studying the Mechanisms of RNA Translocation into Mitochondria
advertisement
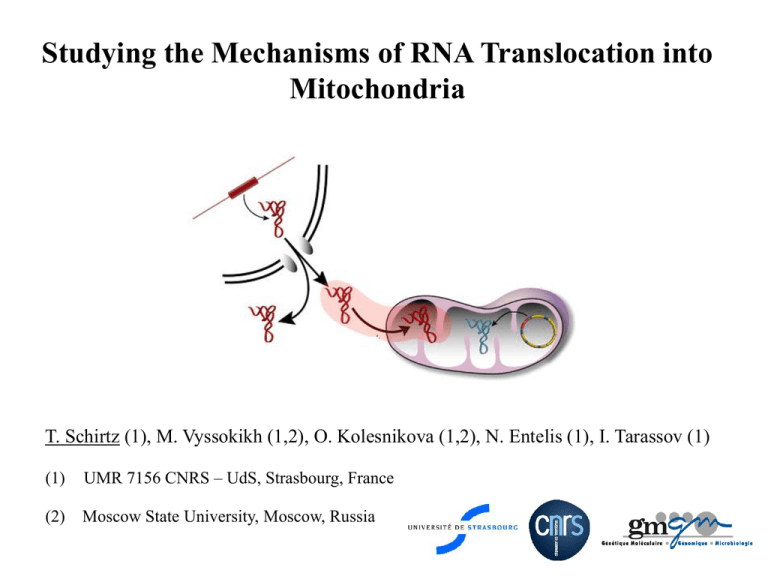
Studying the Mechanisms of RNA Translocation into Mitochondria T. Schirtz (1), M. Vyssokikh (1,2), O. Kolesnikova (1,2), N. Entelis (1), I. Tarassov (1) (1) UMR 7156 CNRS – UdS, Strasbourg, France (2) Moscow State University, Moscow, Russia Mitochondria : the “powerplant” of the cell Fluorescent staining of mitochondrial networks Localization and features - Organelles present in cytoplasm of every eukaryotic cell - Arranged in highly complex networks - Endosymbiotic origin - Possess own genetic information and own genetic code Inner membrane space Electron microscopy Electron microscopy Schematic representation Functions Adenosine tri-phosphate (ATP) INNER MEMBRANE - Respiration and energy production (ATP) ATP - Apoptosis (programmed cell death) - Metabolism (Krebs cycle, urea cycle…) respiratory chain ΔΨ + ΔpH = p.m.f (proton-motive force) - Oxidation and synthesis of fatty acids - Synthesis of essential amino acids - Regulation of intracellular calcium pool -… Mitochondrial RNA import is widespread among species Genus Imported RNA's Number of imported RNA's • Protozoans : (>40 species) tRNA • Plants : (>20 species) tRNA 2 - 15 • Mammals: tRNA 1-2 (4 species) 5S rRNA (MRP RNA, RnaseP RNA) Fungi: (3 species) tRNA 1 - 20 Saccharomyces cerevisiae tRNA 1-3 • majority - totality Why study import of RNA into mitochondria ? Optical nerve atrophy / Retinitis Pigmentosa Respiratory defects Cerebrovascular diseases Mental retardation Cardiomyopathy Liver defects Deafness Human mitochondrial genome Peripherical myopathies Nanisme defects of bone marrow Diabetes Thyroid gland diseases Myopathies Import of tRNALys into mitochondria of Saccharomyces cerevisiae Saccharomyces cerevisiae cytoplasmic (anticodon CUU) cytoplasmic (anticodon UUU) mitochondrial (anticodon UUU) 3 isoforms of tRNALys tRK1 (partially imported) tRK2 (non-imported) tRK3 Cloverleaf structures of cytoplasmic and mitochondrial tRNALys of S.cerevisiae Cytoplasmic targeting factors of tRK1 to the mitochondrial surface Aim : - identification inner of proteins membranes implicated in of outer and of mitochondria the translocation mechanisms of tRNALys (tRK1) - study the electrochemical (ΔΨ) and ATP level (Entelis et al. 2006) Enolase-2 : forms 1st complex with aminoacylated tRK1 Pre-MSK : forms 2nd complex with aminoacylated tRK1 interaction favors complex formation with pre-MSK interaction necessary for tRK1 importation requirements membrane for potential The pre-protein import machinery, porins and import of tRNALys metabolites RNase treatment Incubation 32°C Isolation mitochondrial RNA EDTA washing POR2 Import Mix In vitro import assay - ATP 5mM - NADH 3mM - Succinate 10mM - MgCl2 2,5mM - Sorbitol 0,44M - HEPES-NaOH pH6,8 10mM Exposition photosensitive plate Δ POR 1 Δ TOM 5 Wild type (-) mitocondria 10%PAAG/8M Urea gel electrophoresis Autoradiography Input 5% + RNase Input 5% Δ TIM 44 Δ TOM 20 Δ TOM 70 Wild type Scheme of the pre-protein import machinery (Bolender et al. 2008) Fixation and drying of the gel Δ POR 2 POR1 tRK1 In vitro import of tRK1 into mitochondria of strains carrying deletions for porins and for a non essential protein of the pre-protein import machinery tRK1 % relative import 200 180 160 140 % import In vitro import of tRK1 into mitochondria of strains carrying deletions for essential proteins of the pre-protein import machinery (Tarassov et al. 1995) 120 100 80 60 40 20 0 WT ΔTOM 5 ΔPOR 1 ΔPOR 2 Identification of proteins interacting with tRK1 by North-Western and mass-spectrometry analysis Preparation of mitochondrial outer and inner membranes SDS-Page and transfer to nitrocellulose membrane Renaturation of blotted proteins Probing of the filter with radiolabeled aminoacylated tRK1 Autoradiography Localization of the signals on an identical SDS-Page Wild type ΔPOR1 TOM 5 (component of the translocase of outer membrane complex) + - TOM 20 (component of the translocase of outer membrane complex) - + TOM 22 (component of the translocase of outer membrane complex) - + TOM 40 (component of the translocase of outer membrane complex) - + POR 1 (Porin 1) + - AAC (ADP/ATP carrier) + + MIR 1 (Phosphate carrier) + + QCR 2 (subunit 2 of the ubiquinol cytochrome-c reductase complex) + + PHB 1 (subunit 1 of prohibitin complex) - + TIM 50 (component of the translocase of inner membrane complex) - + Outer membrane: Inner membrane : Identification by nano-LC MS/MS Identification of proteins interacting with tRK1 by crosslinking and SDS-PAGE/Mass-spectrometry analysis Mitoribosomal proteins: MRP-L1 (mitochondrial ribosomal protein of the large subunit) MRP-L3 (mitochondrial ribosomal protein of the large subunit) MRP-L7 (mitochondrial ribosomal protein of the large subunit) MRP-L35 (mitochondrial ribosomal protein of the large subunit) Matrix enzymes: KGD 1 (component of the mitochondrial alpha-ketoglutarate dehydrogenase complex) Mitochondrial chaperones: HSP 60 (mitochondrial chaperonin required for ATP-dependent folding of precursor polypeptides and complex assembly) KGD 2 (dihydrolipoyl transsuccinylase) SHM 1 (mitochondrial serine MIS 1 (mitochondrial C1-tetrahydrofolate synthase) ILV 5 (Acetohydroxyacid reductoisomerase) PDX 1 (lipoamide dehydrogenase) ACO 1 (aconitase) hydroxymethyltransferase) Protein involved in genome maintainance: RIM 1 (single-stranded DNA-binding protein essential for mitochondrial genome maintenance) MSS 116 (DEAD-box protein required for efficient splicing of mitochondrial Group I and II introns) ILV 5 (Acetohydroxyacid reductoisomerase) Dependence of tRK1 import on the level of ATP and electrochemical membrane potential ΔΨ 1 7 8 9 x x x x x x x x x x x x x x x 4 5 x x x x succinate x x x oligomycine x x ATP x 2 6 3 (-) FCCP x DIDS PDE x x x x x x x x x x (+) (+) x x x x tRK1 ATP ATP ATP is more important then electrochemical membrane potential ΔΨ POR1 seems to be implicated in the import mechanism External or internal ATP ? - ATPinternal - + + + + - - - external ATP - + + - - - + + + - + - - + + - + ΔΨ - tRK1 Internal ATP pool is more important than external pool Conclusions and perspectives - ATP is indispensable for import compared to electrochemical membrane potential ΔΨ - Internal ATP pool is more important than external ATP pool - Proteins of the pre-protein import machinery (Tom20, Tom5, Tim44) and porin 1 are implicated in the translocation mechanism - Improve crosslinking approach - Study import in mutants for proteins identified by North-Western and the crosslinkink method - Reconstitution of a minimal import machinery in artificial liposomes Participants and sponsors Team « Mito » O. Kolesnikova O. Karicheva A.-M. Heckel N. Entelis A. Smirnov C. Comte T. Schirtz M. Vyssokikh Y. Tonin R. Martin Y. Kharchenkov I. Tarassov Collaborations: A. Lombès (Salpétrière, Paris) A. Dietrich (IBMP, Strasbourg)