Splicing
advertisement
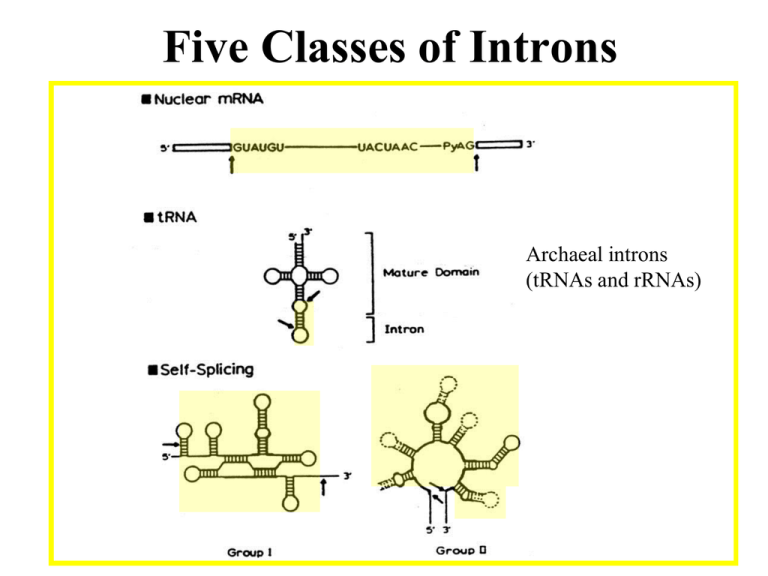
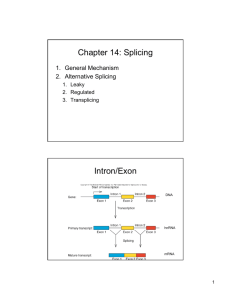
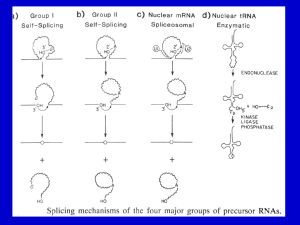
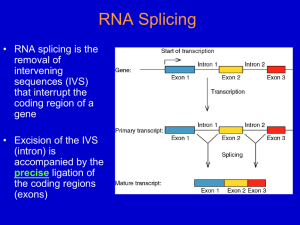
Five Classes of Introns Archaeal introns (tRNAs and rRNAs) Generic Splicing Reaction 5’ splice junction Exon 1 Intron 3’ splice junction Exon 2 Intron Exon 1 Exon 2 Two Steps (“Scissors than Tape”): Step 1: Break phosphodiester bonds at the exon-intron boundaries (splice junctions). 5’ bond broken before 3’ bond Step 2: Formation of a new phosphodiester bond between 3’ end of upstream exon and 5’ end of downstream exon Transesterification • Splicing of Group I, II, and Pre-mRNA introns results from two sequential transesterification reactions • “Transesterification” occurs when a hydroxyl group makes a nucleophilic attack on a phosphodiester bond to form a new phosphodiester bond while displacing a hydroxyl group • The reaction requires no energy (ATP-independent) • Phosphate is conserved Group I Introns Location: • Nuclear rRNA genes of unicellular organisms (e.g. tetrahymena + other ciliates) • Organellar tRNAs and rRNAs (mitochondria and chloroplasts) • rRNA, mRNA, tRNA in bacteria (but rare) • Viruses (e.g. T4 thymidylate synthase mRNA gene) • Not found in vertebrates (e.g. “us”) Role as Mobile Genetic Elements: • introns can encode homing endonucleases that allow intron mobility Group I Intron Structure • Little conservation of primary structure (e.g. P, Q, R, S elements, 3’splice-site G) • All group I introns fold into a characteristic secondary structure (and likely tertiary structure) • X-ray structure has been solved for most of the intron from tetrahymena rRNA • RNA folding is critical for splicing Group I Secondary Structure Internal Guide Sequence G Binding Site (IGS) (Active Site) Conserved G Group I Intron Splicing Mechanism GOH 3’ exon Autocatalytic or “Self-splicing” G 5’ exon intron Step 1 G G Step 2 intron Sequential Transesterfications: Step I: 3’OH of an exogenous guanosine attacks the phosphodiester bond at the 5’ splice site -G covalently linked to intron -5’exon now contains a 3’ OH group Step II: 3’ OH of 5’exon attacks the phosphodiester bond of 3’ splice site -intron is released -exons are ligated together 5’ exon 3’ exon Joined exons (mature RNA) Group II Introns Location: • rRNA, tRNA, mRNA Eukaryotic organelles -mitochondria (fungi), chloroplasts (plants) • mRNA of some Eubacteria (i.e. prokaryotes) Splicing: • Autocatalytic or self-splicing in vitro • proteins required in vivo Role as Mobile Genetic Elements: • Introns often encode reverse transcriptases that allow intron to change genomic position. Structure of Group II Introns • Group II introns exhibit little primary sequence conservation • All fold into a common secondary structure containing six helical domains (d1-d6) that emanate from a “central wheel” • Domains 5 and 6 contain important catalytic activity Tertiary Interactions Critical for Splicing of Group II Introns • Exon binding sequences (EBS 1 and 2) in domain I to intron binding sequences (IBS 1 and 2) near 5’ end of 5’ exon (helps define 5’ splice-site) • Nucleotides in loop of domain 5 interact with nucleotides in domain I • Nucleotides in “wheel” (RGA=g) interact with 3’ splice site (YA= g’) (helps define 3’ splice-site) • Nucleotides in in domain 1(e’) interact with those near 5’ splice site (e) Group II Introns “Catalytic Core” (Active Site) Branch Point Adenosine 5’ EXON 3’ EXON Lariat Intermediate 2’ to 5’ Linkage Splicing Mechanism for Group II and Pre-mRNA Introns 3’ to 5’ Linkage Phosphate is conserved Nuclear Pre-mRNA Introns Location: • Common in vertebrates, numerous introns/gene • Rare in unicellular eukaryotes like yeast, usually one intron/gene when any Conserved Sequences: at splice junctions (GT-AG rule), branch site and polypyrimidine tracts yeast metazoans: 5’ splice site AG/GUAUGU AG/GURAGU branch site polypyrimidine tract UACUAAC Yn YNCURAC YYYYn 3’ splice site CAG/G YAG/G A in branch site adenosine is called the branch point Spacing between the elements is important The 5’ splice site is generally >45 nucleotides from the branch point The 3’ splice site is generally 18-38 nucleotides away from the metazoan branch point and 6-150 nucleotides from the yeast branch point Pre-mRNA Splicing • Requires ~100 proteins and 5 RNAs • Occurs in a large RNP assembly known as the “Spliceosome” • Catalytic component unknown but may be RNA-catalyzed • Splicing via sequential transesterification reactions (same chemical steps as Group II intron splicing) Pre-tRNA Splicing Splice P OH Endonuclease Ligase ‘Kinase’ 3’ phosphodiesterase ‘Cyclic Phosphodiesterase’ ‘Adenylase’ ‘Ligase’ ‘2'-Phospho transferase’ or ATP Splicing of Nuclear Pre-tRNA Introns (in Yeast ) Protein-catalyzed 1) endonuclease 2) ‘ligase’with 5 activities Splicing in Archaea • tRNAs and rRNAs • Endonuclease: -symmetric homodimer - recognizes/cuts a bulge-helix-bulge motif formed by pairing of region near two exonintron junctions • Ligase: - joins exons and circularizes introns Bulge-Helix-Bulge Motif • Two 3 nt bulges on opposite strands separated by 4 bp Buldge Helix Buldge tRNA Processing in Archaea BHB Endonuclease Ligase rRNA Processing in Archaea “Inteins”: Protein Splicing Too! Summary of Intron Splicing Mechanisms Catalytic Mechanisms: nucleophiles, introns, catalysts Splice-site Selection: splice junctions, recognition