Key Concepts
advertisement

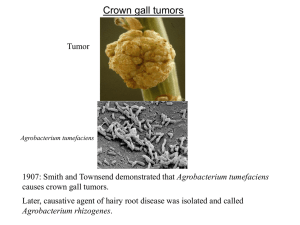
Chapter 16 Extrachromosomal Replication 16.1 Introduction Bacterial extrachromosomal genomes fall into two general types: plasmids and bacteriophages (phages). Some plasmids, and all phages, have the ability to transfer from a donor bacterium to a recipient by an infective process. - plasmids: self-replicating circular molecules of DNA that are maintained in the cell in a stable and characteristic number of copies, that is, the number remains constant from generation to generation. - phages that are found as part of the bacterial chromoosome are aid to show lysogeny; plasmids that have the ability to behave like this are called episomes. 16.2 The Ends of Linear DNA Are a Problem for Replication Key Concepts Special arrangements must be made to replicate the DNA strand with a 5’ end. Figure 16.1: replication of a 5’ end is a problem. Special mechanism must be employed for replication at the ends of linear replicons: - The problem may be circumvented by converting a linear replicon into a circular or multimeric molecule. Phages such as T4 or lambda use such mechanisms (see Section 16.4). - The DNA may form an unusual structure—for example, by creating a hairpin at the terminus, so that there is no free end. - Instead of being precisely determined, the end may be variable. Eukaryotic chromosomes may adopt this solution, in which the number of copies of a short repeating unit at the end of the DNA changes (see Section 28.18). - A protein may intervene to make initiation possible at the actual terminus. Several linear viral nucleic acids have proteins that are covalently linked to the 5’ terminal base. The best characterized examples are adenovirus DNA, phage φ29 DNA, and poliovirus RNA. Figure 16.1. Replication could run off the 3’ end of a newly synthesized linear strand, but could it initiate at a 5’ end? 16.3 Terminal Proteins Enable Initiation at the Ends of Viral DNAs Key Concepts A terminal protein binds to the 5’ end of DNA and provides a cytidine nucleotide with a 3’-OH end that primes replication. Figure 16.2: Adenovirus DNA replicates by strand displacement. In several viruses that use such mechanisms, a protein is found covalently attached to each 5’ end, In the case of adenovirus, a terminal protein is linked to the mature viral DNA via a phosphodiester bond to serine, as indicated in Figure 16.3. The terminal protein carries a cytidine nucleotide that provides the primer, and it is associated with DNA polymerase (Figure 16.4). Figure 16.2. Adenovirus DNA replication is initiated separately at the two ends of the molecule and proceeds by strand displacement. Figure 16.3. The 5’ terminal phosphate at each end of adenovirus DNA is covalently linked to serine in the 55 kD Ad-binding protein. Figure 16.4. Adenovirus terminal protein binds to the 5’ end of DNA and provides a C-OH end to prime synthesis of a new DNA strand. 16.4 Rolling Circles Produce Multimers of a Replicon Key Concepts A rolling circle generates single-stranded multimers of the original sequence. Replication of only one strand is used to generate copies of some circular molecules. A nick opens one strand, and then the free 3’-OH end generated by the nick is extended by the DNA polymerase. Figure 16.5: The rolling circle replicates DNA. Figure 16.6: Rolling circles show in microscopy. Figure 16.7: Rolling circle products are versatile. Figure 16.5. The rolling circle generates a multimeric singlestranded tail. Figure 16.6. A rolling circle appears as a circular molecule with a linear tail by electron microscopy. Figure 16.7. The fate of the displaced tail determines the types of products generated by rolling circles. Cleavage at unit length generates monomers, which can be converted to duplex and circular forms. Cleavage of multimers generates a series of tandemly repeated copies of the original unit. Note that the conversion to doublestranded form could occur earlier, before the tail is cleaved from the rolling circle. 16.5 Rolling Circles Are Used to Replicate Phage Genomes Key Concepts The φX A protein is a cis-acting relaxase that generates singlestranded circles from the tail produced by rolling circle replication. Replication by rolling circles is common among bacteriophages. Unit genomes can be cleaved from the displaced tail, generating monomers that can be packaged into phage particles or used for further replication cycles (Figure 16.8). Phage φX174 consists of a single-stranded circular DNA known as the plus (+) strand. A complementary strand, called the minus (-) strand, is synthesized. This action generates the duplex circle shown at the top of the figure, which is then replicated by a rolling circle mechanism. The A protein (called relaxase) coded by the phage genome nicks the (+) strand of the duplex DNA at a specific site that defines the origin for replication. The structure of the DNA plays an important role in this reaction, for the DNA can be nicked only when it is negatively supercoiled. Figure 16.8. φX174 RF DNA is a template for synthesizing singlestranded viral circles. The A protein remains attached to the same genome through indefinite revolutions, each time nicking the origin on the viral (+) strand and transferring to the new 5’ end. At the same time, the released viral strand is circularized.. 15.6 The F Plasmid Is Transferred by Conjugation between Bacteria Key Concepts A free F factor is a replicon that is maintained at the level of one plasmid per bacterial chromosome. An F factor can integrate into the bacterial chromosome, in which case its own replication system is suppressed. The F factor codes for specific pili that form on the surface of the bacterium. An F pilus enables an F-positive bacterium to contact an F-negative bacterium and to initiate conjugation. Bacterial conjugation: a plasmid genome or host chromosome is transferred from one bacterium to another. Conjugation is mediated by the F plasmid, which is the classic example of an episome—an element that may exist as a free circular plasmid, or that may become integrated into the bacterial chromosome as a linear sequence. The F plasmid is a large circular DNA ~100 kb in length. In its free (plasmid) form, the F plasmid utilizes its own replication origin (oriV) and control system, and is maintained at a level of one copy per bacterial chromosome, this system is suppressed, and F DNA is replicated as a part of the chromosome. A large (~33 kb) region of the F plasmid called the transfer region is required for conjugation. It contains ~40 genes that are required for the transmission of DNA (Figure 16.9). F-positive bacteria possess surface appendages called pili (pilus) that are coded by the F factor. The gene traA codes for the single subunit protein, pilin. Mating is initiated when the tip of the F-pilus contacts the surface of the recipient cell (Figure 16.10). Figure 16.9. The tra region of the F plasmid contains the genes needed for bacterial conjugation. Figure 16.10. Mating bacteria are initially connected when donor F pili contact the recipient bacterium. 16.7 Conjugation Transfers Single-Stranded DNA Key Concepts Transfer of an F factor is initiated when rolling circle replication begins at oriT. The free 5’ end initiates transfer into the recipient bacterium. The transferred DNA is converted into double-stranded form in the recipient bacterium. When an F factor is free, conjugation “infects” the recipient bacterium with a copy of the F factor. When an F factor is integrated, conjugation causes transfer of the bacterial chromosome until the process is interrupted by (random) breakage of the contact between donor and recipient bacteria. Transfer of the F factor is initiated at a site called oriT, the origin of transfer, which is located at one end of the transfer region. Figure 16.11 shows that the freed 5’ end leads the way into the recipient bacterium. When an integrated F plasmid initiates conjugation, the orientation of transfer is directed away from the transfer region and into the bacterial chromosome; such strains are described as Hfr (for high frequency recombination). Figure 16.12 shows that, following a short leading sequence of F DNA, bacterial DNA is transferred. Figure 16.11. Transfer of DNA occurs when the F factor is nicked at oriT and a single strand is led by the 5’ end into the recipient. Only one unit length is transferred. Complementary strands are synthesized to the single strand remaining in the donor and to the strand transferred into the recipient. Figure 16.12. Transfer of chromosomal DNA occurs when an integrated F factor is nicked at oriT. Transfer of DNA starts with a short sequence of F DNA and continues until prevented by loss of contact between the bacteria. 16.8 The Bacterial Ti Plasmid Causes Crown Gall Disease in Plants Key Concepts Infection with the bacterium A. tumefaciens can transform plant cells into tumors. The infectious agent is a plasmid carried by the bacterium. The plasmid also carries genes for synthesizing and metabolizing opines (arginine derivatives) that are used by the tumor cell. Most events in which DNA is rearranged or amplified occur within a genome, but the interaction between bacteria and certain plants involves the transfer of DNA from the bacterial genome to the plant genome. Crown gall disease, shown in Figure 16.13, can be induced in most dicotyledonous plants by the soil bacterium Agrobacterium tumefaciens. The tumor-inducing principle of Agrobacterium resides in the Ti plasmid. Ti plasmids can be divided into four groups, according to the types of opine (novel derivatives of arginine) that are made; Nopaline, Octopine, Agropine, Ri plasmids The types of genes carried by a Ti plasmid are summarized in Figure 16.14. Figure 16.13. An Agrobacterium carrying a Ti plasmid of the nopaline type induces a teratoma, in which differentiated structures develop. Figure 16.14. Ti plasmids carry genes involved in both plant and bacterial functions. 16.9 T-DNA Carries Genes Required for Infection Key Concepts Part of the DNA of the Ti plasmid is transferred to the plant cell nucleus. The vir genes of the Ti plasmid are located outside the transferred region and are required for the transfer process. The vir genes are induced by phenolic compounds released by plants in response to wounding. The membrane protein VirA is autophosphorylated on histidine when it binds an inducer. VirA activates VirG by transferring the phosphate group to it. The VirA-VirG is one of several bacterial two component systems that use a phosphohistidine relay. The interaction between Agrobacterium and a plant cell is illustrated in Figure 16.15. The bacterium does not enter the plant cell, but rather transfers part of the Ti plasmid to the plant nucleus. The transferred part of the Ti genome is called T-DNA. It becomes integrated into the plant genome. Transformation of plant cells requires three types of function carried in the Agrobacterium: - Three loci on the Agrobacterium chromosome, chvA, chvB, and pscA, are required for the initial stage of binding the bacterium to the plant cell. They are responsible for synthesizing a polysaccharide on the bacterial cell surface. - The vir region carried by the Ti plasmid outside the T-DNA region is required to release and initiate transfer of the T-DNA. - The T-DNA is required to transform the plant cell. Figure 16.15. T-DNA is transferred from Agrobacterium carrying a Ti plasmid into a plant cell, where it becomes integrated into the nuclear genome and expresses functions that transform the host cell. The organization of the major two types of Ti plasmid is illustrated in Figure 16.16. The T-region occupies ~23 kb. The virulence genes code for the functions required for the transfer process. Six loci (virA, -B, -C, -D, -E, and –G) reside in a 40-kb region outside the T-DNA. Their organization is summarized in Figure 16.17. We may divide the transforming process into (at least) two stages: - Agrobacterium contacts a plant cell and the vir genes are induced. - vir gene products cause T-DNA to be transferred to the plant cell nucleus, where it is integrated into the genome. Figure 16.18: Wounding produces acetosyringone. Figure 16.19: VIrA-VirG is a two-component system. Figure 16.16. Nopaline and octopine Ti plasmids carry a variety of genes, including T-regions that have overlapping functions. Figure 16.17. The vir region of the Ti plasmid has six loci that are responsible for transferring T-DNA to an infected plant. Figure 16.18. Acetosyringone (4-acetyl-2,6-dimethoxyphenol) is produced by N. tabacum upon wounding and induces transfer of TDNA from Agrobacterium. Figure 16.19. The two-component system of VirA-VirG responds to phenolic signals by activating transcription of target genes. 16.10 Transfer of T-DNA Resembles Bacterial Conjugation Key Concepts T-DNA is generated when a nick at the right boundary creates a primer for synthesis of a new DNA strand. The preexisting single strand that is displaced by the new synthesis is transferred to the plant cell nucleus. Transfer is terminated when DNA synthesis reaches a nick at the left boundary. The T-DNA is transferred as a complex of single-stranded DNA with the VirE2 single strand-binding protein. The single stranded T-DNA is converted into double-stranded DNA and integrated into the plant genome. The mechanism of integration is not known. T-DNA can be used to transfer genes into a plant nucleus. Figure 16.20: T-DNA is bounded by direct repeats. Figure 16.21: A model for transfer. Figure 16.20. T-DNA has almost identical repeats of 25 bp at each end in the Ti plasmid. The right repeat is necessary for transfer and integration to a plant genome. T-DNA that is integrated in a plant genome has a precise junction that retains 1 to 2 bp of the right repeat, but the left junction varies and may be up to 100 bp short of the left repeat. Figure 16.21. T-DNA is generated by displacement when DNA synthesis starts at a nick made at the right repeat. The reaction is terminated by a nick at the left repeat.