Lec. 9
advertisement
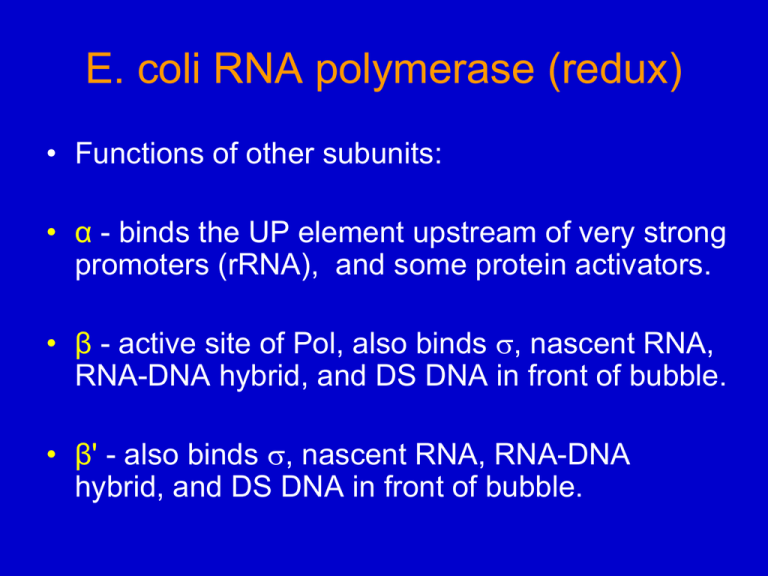
E. coli RNA polymerase (redux) • Functions of other subunits: • α - binds the UP element upstream of very strong promoters (rRNA), and some protein activators. • β - active site of Pol, also binds , nascent RNA, RNA-DNA hybrid, and DS DNA in front of bubble. • β' - also binds , nascent RNA, RNA-DNA hybrid, and DS DNA in front of bubble. Thermus aquaticus RNAP core. “The Claw” From Fig 6.35 T. Aquaticus Holoenzyme Similar to Fig. 6.37 RNAP binds/protects DNA from bp -44 to +3 DNA used to form the RF Complex Fig. 6.40 Locks the enzyme into the open promoter complex. Fig. 6.41a, RF Complex Figure 6.41b, RF with removed Fig 6.43b RF Model with removed. RNAP: Backsliding and Editing 1. If wrong nucleotide incorporated, elongation can become arrested. 2. Backsliding now competes with elongation: – Pol backs up, extruding some of nascent RNA 3. Gre proteins activate RNAP core to cleave small piece that has wrong nucleotide. 4. Pol starts elongating again. RNAP core Square is the next NTP to be added. Green – nascent RNA that will be cleaved off Red – “older RNA” Gene Regulation in Prokaryotes Regulation occurs at every level: 1. Gene organization (operon co-expression) 2. Transcription (repression, activation, attenuation) 3. mRNA stability (affected by translation and the 3’ stem-loops), have “degradosome” 4. Translation (repression, activation and autoregulation) 5. Protein stability and other modifications TRANSCRIPTIONAL CONTROL DOMINANT! Lactose (Lac) Operon • Diauxic growth (2 phases or types, which use different substrates) • Operon organization • Negative and positive regulation – Lac Repressor (lacI gene) – Catabolite Activator Protein (crp gene) Diauxic growth of E. coli on a mixture of lactose + glucose. If E. coli presented with glucose & lactose, use mainly glucose until gone, then use lactose. Lactose Structure & Metabolism CH2OH HO CH2OH O OH l ac t ose -galactosidase O OH O OH OH HO galactose glucose Glucose Galact ose lactose ep im erase g ly co ly sis Lac Operon: Repression Figure 7.3 Inducer : Allolactose, produced by lacZ Fig. 7.4 A side reaction of lacZ. - 1,6 linked 1965 Nobel Prize in Physiology or Medicine (for their work on the lac operon and bacterial genetics) J. Monod F. Jacob A. Lwoff Equilibrium DNA – Protein Binding Example: lacI + DNAo ↔ lacI-DNAo lacI – lac repressor DNAo – lac operator DNA Kd = [lacI] [DNAo] ∕ [lacI-DNAo] Kd = equilibrium dissociation constant 1 x 10-8 to 10-12 M = high affinity Figure 7.6 Lac repressor binding to lac operator. IPTG = synthetic inducer of lac operon. Figure 7.7 There are really 3 operator regions for the Lac Operon. CAP – activator protein RNAP – RNA polymerase Fig 7.10 Operators work cooperatively (synergistically). DNAs introduced into E. coli genome of a lacZ mutant using λ phage. LacI gene was present in the chromosome. IPTG was used to induce lacZ. Numbers are based on the ratio of lacZ activity in the presence and absence of inducer (IPTG). Structural basis for cooperativity of operators: Lac repressor can bind 2 operator sequences. DNA LacI Tetramer (2 dimers held together at the bottom) Fig 7.12a







![Lac Operon AP Biology PhET Simulation[1]](http://s3.studylib.net/store/data/006805976_1-a15f6d5ce2299a278136113aece5b534-300x300.png)