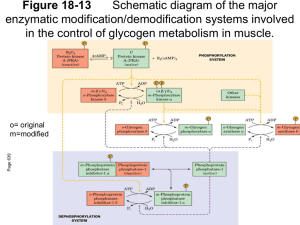
Enzyme Regulatory Strategies
advertisement

Enzyme Regulation What you Need to Know • Understand the different ways that enzyme activity is regulated. • What factors influence enzyme activity? • General understanding of symmetry model versus sequential model of allosteric regulation • How do positive versus negative effectors affect plots of enzyme velocity versus substrate concentration? • What are general classes of protein kinases? • How is cAMP protein kinase regulated? • Understand the different ways that Glycogen phosphorylase is regulated What Factors Influence Enzymatic Activity? • The availability of substrates and cofactors usually determines how fast the reaction goes • As product accumulates, the apparent rate of the enzymatic reaction will decrease • Genetic regulation of enzyme synthesis and decay determines the amount of enzyme present at any moment • Enzyme activity can be regulated allosterically (instantaneous response) • Enzyme activity can be regulated through covalent modification (interconvertable enzymes) (response times of seconds or less) – i.e. protein kinases (activate Ser, Thr, Tyr side chains) • Zymogens (irreversible process), isozymes, and modulator proteins may play a role Transcription Regulation • The amount of enzyme synthesized by a cell is determined by transcription regulation. – Induction – Repression • Genetic controls over enzyme levels have a response time of minutes to hours or longer. • Once synthesized, an enzyme can be degraded via normal turnover of the protein or through specific decay mechanisms that target the enzyme for destruction (i.e. ubiquitinin pathway). What Factors Influence Enzymatic Activity? Enzyme regulation by reversible covalent modification. What Factors Influence Enzymatic Activity? Zymogens are inactive precursors of enzymes. Typically, proteolytic cleavage produces the active enzyme. Proinsulin is an 86-residue precursor to insulin The proteolytic activation of chymotrypsinogen Proteolytic Enzymes of the Digestive Tract The Cascade of Activation Steps Leading to Blood Clotting The intrinsic and extrinsic pathways converge at factor X, and the final common pathway involves the activation of thrombin (cleaves at ArgGly peptide) and its conversion of fibrinogen into fibrin, which aggregates into ordered filamentous arrays that become crosslinked to form the clot. Isozymes Are Enzymes With Slightly Different Subunits (structurally similar but catalytically distinct) Muscle becomes anaerobic: pyruvate from glucose via glycolysis. Requires LDH to regenerate NAD+ so glycolysis can continue. (A4 best at regenerating NAD+) Heart is aerobic using lactate as fuel converting it to pyruvate; to fuel Citric Acid Cycle (B4 inhibited by excess pyruvate) Isozymes of lactate dehydrogenase (LDH). What Are the General Features of Allosteric Regulation? Action at "another site" • Enzymes situated at key steps in metabolic pathways are modulated by allosteric effectors • These effectors are usually produced elsewhere in the pathway • Effectors may be feed-forward activators or feedback inhibitors – Sigmoid Kinetics ("S-shaped") • Do not obey Michaelis-Menten eq. • Suggest 2nd order or higher relationship between v and [S] v proportional to [S]n – Explanation: Cooperative Binding Sigmoid v versus [S] plot. The dotted line represents the hyperbolic plot characteristic of normal Michaelis-Menten kinetics. Can Allosteric Regulation Be Explained by Conformational Changes in Proteins? • Monod, Wyman, Changeux (MWC) Model: allosteric proteins can exist in two states: R (relaxed) and T (taut) • In this model, all the subunits of an oligomer must be in the same state • T state predominates in the absence of substrate S • S binds much tighter to R than to T and substrate inhibitors bind only to the T state The Symmetry Model for Allosteric Regulation is Based on Two Conformational States for a Protein Allosteric effects: A and I binding to R and T, respectively. The Symmetry Model for Allosteric Regulation is Based on Two Conformational States for a Protein Allosteric effects: A and I binding to R and T, respectively. The Symmetry Model for Allosteric Regulation is Based on Two Conformational States for a Protein Allosteric effects: A and I binding to R and T, respectively. More about the MWC model • Cooperativity is achieved because S binding increases the population of R, which increases the sites available to S • Ligands such as S are positive homotropic effectors • Molecules that influence the binding of something other than themselves are heterotropic effectors The Sequential Model for Allosteric Regulation is Based on Ligand-Induced Conformation Changes • An alternative model – proposed by Koshland, Nemethy, and Filmer (the KNF model): ligand binding triggers a conformation change in a protein • If the protein is oligomeric, ligand-induced conformation changes in one subunit may lead to conformation changes in adjacent subunits • The KNF model explains negative cooperativity • The KNF model is termed the sequential model The Sequential Model for Allosteric Regulation is Based on Ligand-Induced Conformation Changes The Sequential Model for Allosteric Regulation is Based on Ligand-Induced Conformation Changes The KoshlandNemethy-Filmer model. Theoretical curves for the binding of a ligand to a protein having four identical subunits, each with one binding site for the ligand. Novel Activation and Regulation • Pro-enzyme: Inactive with conventional catalytic triad and deformed substrate binding pocket. • Resting enzyme: Inactive with mis-oriented catalytic triad and fully formed substrate binding pocket. • Active enzyme: Fully active, with conventional catalytic triad and fully formed substrate binding pocket. Factor D Pro-enzyme Activated Enzyme Factor D Trypsin Covalent Modification Regulates the Activity of Enzymes • Enzyme activity can be regulated through reversible phosphorylation – This is the most prominent form of covalent modification in cellular regulation • Phosphorylation is accomplished by protein kinases – Each protein kinase targets specific proteins for phosphorylation • Phosphoprotein phosphatases catalyze the reverse reaction – removing phosphoryl groups from proteins • Kinases and phosphatases themselves are targets of regulation Protein Kinases • Protein kinases phosphorylate Ser, Thr, and Tyr residues in target proteins • Kinases typically recognize specific amino acid sequences in their targets (i.e. PKA phosphorylated proteins having searing or threonine residues within an R(R/K)X(S/T) target consensus sequence • In spite of this specificity, all kinases share a common catalytic mechanism based on a conserved core kinase domain of about 260 residues • Kinases are often regulated by intrasteric control, in which a regulatory subunit (or domain) has a pseudosubstrate sequence that mimics the target sequence, minus the phosphorylatable residue (i.e. RRGAI) – A is stericly similar to S but lacks a phosphorylatable OH group. Protein Kinases (a protein Superfamily) Protein kinase A is shown complexed with a pseudosubstrate peptide (orange). This complex also includes ATP (red) and two Mn2+ ions (yellow) bound at the active site. Protein Kinases Phosphorylation is Not the Only Form of Covalent Modification that Regulates Protein Function Cyclic AMP-dependent protein kinase is composed of catalytic and regulatory subunits cyclic AMP-dependent protein kinase (also known as protein kinase A (PKA) is a 150- to 170-kD R2C2 tetramer in mammalian cells. The two R (regulatory) subunits bind cAMP; cAMP binding releases the R subunits from the C (catalytic) subunits. C subunits are enzymatically active as monomers. Some Enzymes are Controlled by Both Allosteric Regulation and Covalent Modification • Glycogen phosphorylase (GP) is an example of the many enzymes that are regulated both by allosteric controls and by covalent modification • GP cleaves glucose units from nonreducing ends of glycogen (phosphorolysis reaction) • This converts glycogen into readily usable fuel in the form of glucose-1-phosphate GP converts glycogen into glucose-1-phosphate • Glycogen phosphorylase (GP) is regulated both by allosteric controls and by covalent modification Phosphoglucomutase converts glucose-1-P into the glycolytic substrate, glucose-6-P The phosphoglucomutase reaction. The structure of glycogen phosphorylase • Glycogen phosphorylase is a dimer of identical 842 residue subunits • Each subunit contains an active site (at the center of the subunit) and an allosteric effector site near the subunit interface • A regulatory phosphorylation site is located at Ser14 on each subunit • A glycogen-binding site exerts regulatory control • Each subunit contributes a “tower helix” (residues 262 to 278) to the subunit-subunit interface • In the dimer, the tower helices extend from their respective subunits and pack against each other The structure of glycogen phosphorylase Glycogen Phosphorylase Activity is Regulated Allosterically • Muscle glycogen phosphorylase shows cooperativity in substrate binding • ATP and glucose-6-P are allosteric inhibitors of glycogen phosphorylase • AMP is an allosteric activator of glycogen phosphorylase • When ATP and glucose-6-P are abundant, glycogen breakdown is inhibited • When cellular energy reserves are low (i.e., high [AMP] and low [ATP] and [G-6-P]) glycogen catabolism is stimulated Glycogen Phosphorylase Activity is Regulated Allosterically v versus S curves for glycogen phosphorylase. (a) The response to the concentration of the substrate phosphate (Pi). (b) ATP is a feedback inhibitor. (c) AMP is a positive effector. It binds at the same site as ATP providing reciprocal regulation. Glycogen phosphorylase conforms to the MWC model • • • • The active form of the enzyme is designated the R state The inactive form of the enzyme is denoted the T state AMP promotes the conversion to the active state ATP, glucose-6-P, and caffeine favor conversion to the inactive T state • A significant conformation change occurs at the subunit interface between the T and R state • This conformational change at the interface is linked to a structural change at the active site that affects catalysis Glycogen Phosphorylase is Controlled by Both Allosteric Regulation and Covalent Modification The mechanism of covalent modification and allosteric regulation of glycogen phosphorylase. A Conformation Change Regulates Activity of Glycogen Phosphorylase The major conformational change that occurs in the Nterminal residues upon phosphorylation of Ser14. Ser14 is shown in red. N-terminal conformation of phosphorylated enzyme (phosphorylase a): yellow. N-terminal conformation of unphosphorylated enzyme (phosphorylase b): cyan. Regulation of GP by Covalent Modification • In 1956, Edwin Krebs and Edmond Fischer showed that a ‘converting enzyme’ could convert phosphorylase b to phosphorylase a • Three years later, Krebs and Fischer show that this conversion involves covalent phosphorylation • This phosphorylation is mediated by an enzyme cascade Glycogen phosphoryase is activated by a cascade of reactions The hormone-activated enzymatic cascade that leads to activation of glycogen phosphorylase. cAMP is a Second Messenger • Cyclic AMP is the intracellular agent of extracellular hormones - thus a ‘second messenger’ • Hormone binding stimulates a GTP-binding protein (G protein), releasing G(GTP) • Binding of G(GTP) stimulates adenylyl cyclase to make cAMP Enzyme Cascade Regulates Phosphorylase Covalent Modification Receptor Cell Membrane Gα (GTP) Gα (GTP) Adenylyl Cyclase Hormone cAMP cAMP G-protein (GDP) GDP GTP active Glycogen Pa 2ADP 2ATP ATP cAMP Inactive cAMP-DPK P P active PK Inactive Glycogen Pb cAMP ADP ATP Inactive PK Active cAMP-DPK