Slides
advertisement

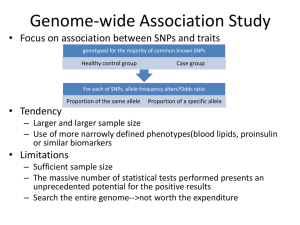
Are you ready for the genomic age? An introduction to human genomics Jacques Fellay EPFL School of Life Sciences Swiss Institute of Bioinformatics Lausanne, Switzerland What is the genome? “It's a shop manual, with an incredibly detailed blueprint for building every human cell. It's a history book - a narrative of the journey of our species through time. It's a transformative textbook of medicine, with insights that will give health care providers new powers to treat, prevent and cure disease.” Francis Collins Glossary • Genome: the complete genetic constitution of an organism, encoded in nucleic acids • Gene: discrete DNA sequence encoding a protein The human genome 3 billions base pairs (ATGC) 20’000 protein-coding genes 99.6% inter-individual identity (yet 4 millions differences) 99% identical to chimpanzee genome (yet 6% different genes) 2001: A Species Odyssey Exploring the human genome 2002 Sanger sequencing, targeted genotyping 2008 Genome-wide genotyping (GWAS) Exome Genome sequencing sequencing International HapMap Project Identification of common genetic variation in 270 individuals from 4 populations • CEU: CEPH (Utah residents with ancestry from northern and western Europe) (30 trios) • CHB: Han Chinese in Beijing, China (45 individuals) • JPT: Japanese in Tokyo, Japan (45 individuals) • YRI: Yoruba in Ibadan, Nigeria (30 trios) 1000 Genomes Project Whole genome sequencing and complete description of human genetic diversity in >1000 individuals from multiple world populations www.1000genomes.org Short video – Sequencing the genome http://ed.ted.com/lessons/how-to-sequence-thehuman-genome-mark-j-kiel We are all different… 4 million DNA variants / individual Single nucleotide variants Multi-nucleotide variants • Small insertions/deletions (indels) • Large copy number variants (CNVs) • Inversions • Translocations • Aneuploidy Glossary • SNV = single nucleotide variant: DNA sequence variation in which a single nucleotide — A, T, C or G — differs between members of the same species • SNP = single nucleotide polymorphism: SNV occurring commonly within a population (> 1%) SNV/SNP Glossary • Allele: One of a number of alternative forms of the same genetic locus (for example a SNP) About 2% of people have two copies of the APOE4 allele and are very likely to succumb to Alzheimer’s disease About 1% of us have two copies of a small deletion in CCR5 and are largely immune to infection by the HIV virus And about 7% do not make any functional CYP2D6 enzyme and therefore codeine provides no pain relief Glossary • Linkage Disequilibrium (LD): Non-random association of alleles that descend from single, ancestral chromosomes (i.e. usually close to each other) • Haplotype: Combination of alleles at adjacent locations on a chromosome that are inherited together How to read the genome? Genotyping Sequencing Glossary • Genotyping: Process of determining genetic differences between individuals by using a set of markers • Sequencing: Process of determining the full nucleotide order of a DNA sequence Genotyping Genome-wide chips: 500K to >1 mio single nucleotide polymorphisms (SNPs) SNP output rs1372493 rs1372493 1.60 16000 1.40 14000 1.20 12000 1 8000 Norm R Intensity (B) 10000 6000 0.80 0.60 4000 0.40 2000 0.20 0 0 -2000 2317 834 74 -0.20 0 2000 4000 6000 8000 10000 Intensity (A) 12000 14000 16000 18000 20000 0 0.20 0.40 0.60 Norm Theta 0.80 1 Homozygous 1 Heterozygous Homozygous 2 Allele frequency of variant <<<<<1% >5% Sequencing +++ Genome-wide genotyping ++ Clinical impact + High-throughput Sequencing (NGS) – Huge amount of data (terabytes) – Analysis computationally intensive – Dedicated IT infrastructure • Pipeline FastQ format – single read @G:1:1:11:1079#0/1 TGATTGATTCCATTCCATTCCATTCCATTTCATTCCATTGCAATCCCTTCCAATCCATTCCATTCCATTCCATTC +G:1:1:11:1079#0/1 `Xa^YO\_^a_`__`a__^a^a^_a``^_\`\\]``[XUGXXXXXWUTWWVWUSTXXPUWYYRVWYYYXZYXYWZ A complete, high-coverage genome will have over 1 billion reads • Pipeline • Pipeline • Pipeline • Pipeline http://www.ncbi.nlm.nih.gov/core/assets/variation/images/popfreq_example.jpg • Pipeline Summary of a single human genome SNVs Premature stop 3.5 million 80 Stop loss Non-synonymous Synonymous 10 11,000 11,000 Essential splice site 25 indels 300,000 Frameshift In-frame 80 200 Whole genome vs. exome sequencing Exome -Coding regions -Cheaper/Faster -Uneven capture of both alleles -Incomplete capture of target region -Bias towards known biology Genome -Complete sequence -Expensive/Throughput -IT issues Clinical sequencing? “Sequencing of the genome or exome for clinical applications has now entered medical practice. Several thousand tests have already been ordered for patients, with the goal of establishing diagnoses for rare, clinically unrecognizable, or puzzling disorders that are suspected to be genetic in origin.” Leslie G. Biesecker and Robert C. Green, NEJM, 19 June 2014 Clinical sequencing? TODAY • Rare functional variants (Mendelian diseases) • Pharmacogenetic variants (150 gene-drug pairs in the FDA “Table of Pharmacogenomic Biomarkers in Drug Labels”, but only 40 genes involved) • Oncogenomics IL28B genotype and response to anti-hepatitis C treatment Ge, Fellay et al. Nature 2009 Clinical sequencing? TOMORROW • Neonatal sequencing • Maternal blood sequencing • DTC genomics brought to doctors Clinical sequencing? LATER • Complex trait genomics (genome data in every health record) – will depend on indepth understanding of functional genomic variation A revolution in the making Eric Green et al., Charting a course for genomic medicine from base pairs to bedside, Nature 2011 Perspective • Genomic-based medicine is around the corner • Considerable space for new (personal) genomic market in health, nutrition, well-being… • Genomic-based medicine is only the beginning of “big-data-based” personalized healthcare Perspective None of this can happen without trust