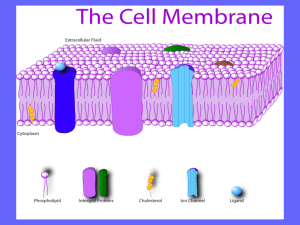
Powerpoint File
advertisement
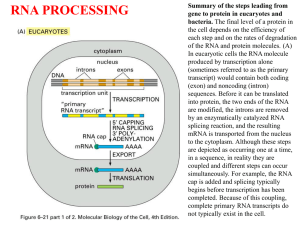
Chapter 1 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Human Physiology Physiology: study of how body works to maintain life Pathophysiology: how physiological processes are altered in disease or injury 1-3 Homeostasis 1-9 Homeostasis Is maintenance of a state of dynamic constancy In which conditions are stabilized above and below a physiological set point By negative feedback loops 1-10 Negative Feedback Loops Sensor: Detects deviation from set point Integrating center: Determines response Effector: Produces response 1-11 Homeostasis continued Regulatory mechanisms: Intrinsic control is built into organ being regulated 1-12 Homeostasis continued Regulatory mechanisms: Extrinsic control comes from outside of organ E.g. body temperature is controlled by antagonistic effects of sweating and shivering 1-13 Homeostasis continued Regulatory mechanisms: Positive feedback is rare because it amplifies changes It is involved in producing blood clots In females it is used to create the LH surge that causes ovulation Positive feedback between the uterus and oxytocin secretion occurs during labor 1-15 Negative Feedback Hormonal Control of Blood Glucose 1-17 Chapter 3 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Gene Expression 3-26 Gene Expression Genes are lengths of DNA that code for synthesis of RNA mRNA carries info for how to make a protein Is transported out of nucleus to ribosomes where proteins are made 3-27 Gene Expression continued Takes place in 2 stages: Transcription occurs when DNA sequence in a gene is turned into a mRNA sequence Translation occurs when mRNA sequence is used to make a protein 3-28 Gene Expression continued Each nucleus contains 1 or more dark areas called nucleoli These contain genes actively making rRNA 3-29 Genome and Proteome Genome refers to all genes in an individual or in a species Proteome refers to all proteins produced by a genome 3-30 Chromatin Is made of DNA and its associated proteins (=histones) Histones are positively charged and form spools around which negatively charged DNA strands wrap Each spool and its DNA is called a nucleosome 3-31 Chromatin continued Euchromatin is the part of chromosomes active in transcription Light in color Heterochromatin is highly condensed region where genes are permanently inactivated 3-32 Chromatin continued 3-33 RNA Synthesis One gene is several thousand nucleotide pairs long DNA in a human cell contains over 3 billion base pairs This is enough to code for at least 3 million proteins But, only a fraction of DNA used for proteins Rest is redundant or may be inactive 3-34 RNA Synthesis continued For transcription, RNA polymerase binds to a “start” sequence on DNA and unzips strands Nearby are promoter regions, which regulate levels of transcription Transcription factors must bind to promoter to initiate transcription 3-35 RNA Synthesis continued Only one strand of DNA contains the gene and is transcribed Its bases pair with complementary RNA bases to make mRNA G pairs with C A pairs with U RNA polymerase detaches when hits a "stop" sequence 3-36 RNA Synthesis continued Transcription produces four types of RNA: pre-mRNA - altered in nucleus to form mRNA mRNA - contains the code for synthesis of a protein tRNA (transfer RNA) - decodes the info contained in mRNA rRNA - forms part of ribosomes 3-37 RNA Synthesis continued Pre-mRNA is much larger than mRNA Contains non-coding regions called introns Coding regions are called exons In nucleus, introns are removed and ends of exons spliced together to produce final mRNA 3-38 RNA Synthesis continued Human genome has <25,000 genes Yet produces >100,000 different proteins 1 gene codes for an average of 3 different proteins Accomplished by alternative splicing of exons This allows a given gene to produce several different mRNAs 3-39 RNA Synthesis continued A newly discovered type of RNA is involved in regulating gene expression These perform RNA interference (RNAi) or silencing Interfere with or silence expression of some genes siRNA (short interfering RNA) and miRNA (micro RNA) molecules pair in varying degrees with different mRNAs Thereby interfering with expression of those mRNAs 1 miRNA may interfere with up to 200 different mRNAs 3-40 Protein Synthesis Occurs one amino acid at a time according to sequence of base triplets in mRNA In cytoplasm, mRNA attaches to ribosomes forming a polysome where translation occurs 3-41 Protein Synthesis continued Ribosomes read 3 mRNA bases (= a triplet) at a time Each triplet is a codon, which specifies an amino acid Ribosomes translate codons into an amino acid sequence that becomes a polypeptide chain 3-42 Protein Synthesis continued 3-43 Protein Synthesis continued Translation of codons is achieved by tRNA and enzymes tRNA contains 3 loops, one of which contains an anticodon Which is complementary to a specific mRNA codon Each tRNA carries the amino acid specified by its anticodon 3-44 Protein Synthesis continued In a ribosome, anticodons on tRNA bind to mRNA codons Amino acids on adjacent tRNAs are brought together and linked enzymatically by peptide bonds This forms a polypeptide; at a stop codon it detaches from ribosome 3-45 Functions of ER Proteins to be secreted are made in ribosomes of rough ER Contain a leader sequence of 30+ hydrophobic amino acids that directs such proteins to enter cisternae of ER Where leader sequence is removed; protein is modified 3-46 Functions of Golgi Secretory proteins leave ER in vesicles and go to Golgi In the Golgi complex carbohydrates are added to make glycoproteins Vesicles leave Golgi for lysosomes or exocytosis 3-47 Protein Degradation The activity of many enzymes and regulatory proteins is controlled by rapidly degrading them By proteases in lysosomes And by cytoplasmic proteasomes 3-48 Chapter 6 Copyright © The McGraw-Hill Companies, Inc. Permission required for reproduction or display. Transport Across Plasma Membrane Plasma membrane is selectively permeable-allows only certain kinds of molecules to pass Many important molecules have transporters and channels Carrier-mediated transport involves specific protein transporters Non-carrier mediated transport occurs by diffusion 6-7 Transport Across Plasma Membrane continued Passive transport moves compounds down concentration gradient; requires no energy Active transport moves compounds against a concentration gradient; requires energy and transporters 6-8 Diffusion Is random motion of molecules Net movement is from region of high to low concentration 6-9 Diffusion continued Non-polar compounds readily diffuse thru cell membrane Also some small molecules such as CO2 and H 2O Gas exchange occurs this way 6-10 Diffusion continued Cell membranes are impermeable to charged and most polar compounds Charged molecules must have an ion channel or transporter to move across membrane 6-11 Diffusion continued Rate of diffusion of a compound depends on: Magnitude of its concentration gradient Permeability of membrane to it Temperature Surface area of membrane 6-12 Osmosis 6-13 Osmosis Is net diffusion of H2O across a selectively permeable membrane H2O diffuses down its concentration gradient H2O is less concentrated where there are more solutes Solutes have to be osmotically active i.e., cannot freely move across membrane 6-14 Osmosis continued H2O diffuses down its concentration gradient until its concentration is equal on both sides of a membrane Some cells have water channels (aquaporins) to facilitate osmosis 6-15 Osmotic Pressure Is the force that would have to be exerted to stop osmosis Indicates how strongly H2O wants to diffuse Is proportional to solute concentration 6-16 Molarity and Molality 1 molar solution (1.0M) = 1 mole of solute dissolved in 1L of solution Doesn't specify exact amount of H2O 1 molal solution (1.0m) = 1 mole of solute dissolved in 1 kg H2O 6-17 Molarity and Molality continued Osmolality (Osm) is total molality of a solution e.g., 1.0m of NaCl yields a 2 Osm solution Because NaCl dissociates into Na+ and Cl- 6-18 Tonicity Is the effect of a solution on osmotic movement of H2O Isotonic solutions have same osmotic pressure Hypertonic solutions have higher osmotic pressure and are osmotically active Hypotonics have lower osmotic pressure Isosmotic solutions have same osmolality as plasma Hypo-osmotic solutions have lower osmotic pressure than plasma Hyperosmotics have higher pressure than plasma 6-19 6-20 Regulation of Blood Osmolality Blood osmolality is maintained in narrow range around 300mOsm If dehydration occurs, osmoreceptors in hypothalamus stimulate: ADH release Which causes kidney to conserve H2O and thirst 6-21 Membrane Transport Systems 6-22 Carrier-Mediated Transport Molecules too large and polar to diffuse are transported across membrane by protein carriers 6-23 Carrier-Mediated Transport continued Protein carriers exhibit: Specificity for single molecule Competition among substrates for transport Saturation when all carriers are occupied This is called Tm (transport maximum) 6-24 Facilitated Diffusion Is passive transport down concentration gradient by carrier proteins 6-25 An Active Transport Pump Is transport of molecules against a concentration gradient ATP is required A carrier protein is required 6-26 Na+/K+ Pump Uses ATP to move 3 Na+ out and 2 K+ in Against their gradients 6-27 Secondary Active Transport ATP needed for “uphill” (against the concentration gradient) movement of molecule or ion obtained from “downhill” (with the concentration gradient) transport of Na+ into cell 6-28 Secondary Active Transport continued Cotransport (symport) is secondary transport in same direction as Na+ Countertransport (antiport) moves molecule in opposite direction to Na+ 6-29 Transport Across Epithelial Membranes Absorption is transport of digestion products across intestinal epithelium into blood Reabsorption transports compounds out of urinary filtrate back into blood 6-30 Transport Across Epithelial Membranes continued Transcellular transport moves material from 1 side to other of epithelial cells Paracellular transport moves material through tiny spaces between epithelial cells 6-31 Transport Across Epithelial Membranes continued Transport between cells is limited by junctional complexes that connect adjacent epithelial cells 6-32 Transport Across Epithelial Membranes continued Plasma membranes can join together to form tight junctions In adherens junctions membranes are “glued” together by proteins that pass through both membranes and attach to cytoskeletons In desmosomes proteins “button” two membranes together 6-33 Bulk Transport Moves large molecules and particles across plasma membrane Occurs by endocytosis and exocytosis (Ch 3) 6-34 Membrane Potential 6-35 Membrane Potential Is difference in charge across membranes Results in part from presence of large anions being trapped inside cell Diffusable cations such as K+ are attracted into cell by anions Na+ is not permeable and is actively transported out 6-36 Equilibrium Potential Describes voltage across cell membrane if only 1 ion could diffuse If membrane permeable only to K+, it would diffuse until it reaches its equilibrium potential (Ek) K+ is attracted inside by trapped anions but also driven out by its concentration gradient At K+ equilibrium, electrical and diffusion forces are = and opposite Inside of cell has a negative charge of about 90mV 6-37 Nernst Equation (Ex) Gives membrane voltage needed to counteract concentration forces acting on an ion Value of Ex depends on ratio of ion concentrations inside and outside cell membrane Ex = 61 log [Xout] z [Xin] Z = valence of the ion 6-38 Nernst Equation (Ex) continued Ex = 61 log [Xout] z [Xin] For concentrations shown at right: Calculate EK+ Calculate ENa+ 6-39 Nernst Equation (Ex) continued EK+ = 61 log 5 +1 150 = -90mV ENa+ = 61 log 145 +1 12 = +66mV 6-40 Resting Membrane Potential (RMP) Is membrane voltage of cell not producing impulses RMP of most cells is –65 to –85 mV RMP depends on concentrations of ions inside and out And on permeability of each ion Affected most by K+ because it is most permeable 6-41 Resting Membrane Potential (RMP) continued Na+ diffuses in so RMP is less negative than EK+ Some 6-42 Role of Na+/K+ Pumps in RMP Because 3 Na+ are pumped out for every 2 K+ taken in, pump is electrogenic It adds about –3mV to RMP 6-43 Summary of Processes that Affect the Resting Membrane Potential 6-44