Lecture 12 - Computational & Systems Biology
advertisement
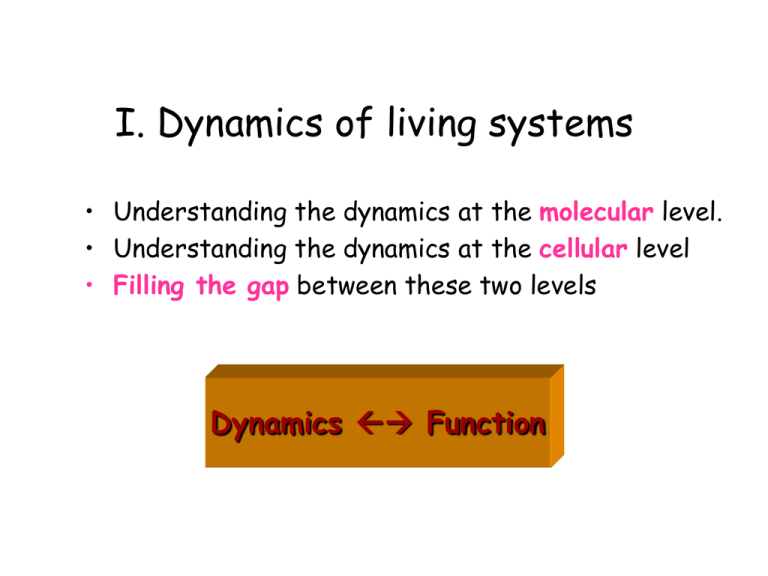
I. Dynamics of living systems • Understanding the dynamics at the molecular level. • Understanding the dynamics at the cellular level • Filling the gap between these two levels Dynamics Function Life’s complexity pyramid Oltvai & Barabasi, Science 2002, 298, 763-764 “The complexity pyramid might not be specific only to cells” Increasing specificity/chemistry) Dominance of molecular machinery Different levels of structural organization: microtubules Challenge: to understand the long-time dynamics of large systems Model: Coarse-grained Method: Analysis of principal modes of motion (Frame transformation: Cartesian collective coordinates) What is the optimal (realistic, but computationally efficient) model for a given scale (length and time) of representation? Which level of details is needed for representing global (collective) motions? How much specificity we need for modeling large scale systems and/or motions? What should be the minimal ingredients of a simplified (reductionist) model? Protein dynamics C (a) Passage over one or more energy barriers Transitions between infinitely many conformations Fluctuations near the folded state Local conformational changes Fluctuations near a global minimum (b) Energy Folding/unfolding dynamics U N' N Conformational space coordinate Can we predict fluctuations dynamics from native state topology only? Gaussian Network Model FOR MORE INFO… Bahar, I., Atilgan, A.R., & Erman, B. (1997) Folding & Des. 2, 173. Flory, P.J. (1976) Proc. Roy. Soc. London A. 351, 351. Detailed specific potentials Approximate uniform potential “A single parameter potential is sufficient to reproduce the slow dynamics in good detail” Rouse chain Connectivity matrix 1 -1 = -1 2 -1 -1 2 -1 .. ... -1 Vtot = (g/2) [ (DR12)2 + (DR23)2 + ........ (DRN-1,N)2 ] = (g/2) [ (DR1 - DR2)2 + (DR2 - DR3)2 + ........ 2 -1 -1 1 Kirchhoff matrix of contacts = 1 if rik < rcut ik= 0 if rik > rcut ii = - Sk ik Vtot = (g/2) DRT DR Comparison with X-ray Temperature Factors 100 (b) 1omf theory 75 Debye-Waller factors: Bk = 8 2 <DRk DRk> /3 experiments 50 25 3 0 0 50 100 150 200 250 300 80 (a) 2ccya 60 40 20 0 0 20 40 60 80 100 FOR MORE INFO... Bahar, I., Atilgan, A.R., & Erman, B. (1997) Folding & Design 2, 173-181 120 350 Comparison with H/D Exchange – NMR data DSi = k ln W(DRi) = - g (DRi)2/ (2T [-1]ii) 20 BPTI 15 Free energy change/RT 10 5 0 0 10 20 30 40 50 60 SNase 12 8 4 0 0 20 40 60 80 100 120 140 residue Bahar, I., Wallquist, A., Covell, D.G., and Jernigan, R.L. (1998) Biochemistry 37, 1067. Covariance matrix (directly found from MD or MC trajectories) <DR1 . DR1> <DR1 . DR2> <DR2. DR1> <DR2 . DR2> <DR1 . DR3> C= <DRN . DRN> DRi = instantaneous fluctuation in the position vector Ri of atom i= Ri - <Ri> <DR1 . DR1> = ms fluctuation of site 1 averaged over all snapshots. Eigenvalue decomposition of C C = U L U-1 U is the matrix of eigenvectors, L is the diagonal matrix of eigenvalues. The ith column (eigenvector) of U is given by a linear combination of Cartesian coordinates and represents the axis of the ith collective coordinate (principal axis) in the conformational space. The ith eigenvalue represents the mean-square fluctuation along the ith principal axis. The motion along the ith principal axis is the ith mode. Decomposition into normal modes • Slowest (global) modes function • Fastest (local) modes stability FOR MORE INFO... Bahar, I., Atilgan, AR, Demirel MC, Erman B. (1998) Physical Review Lett. 80, 2733. Demirel MC, Atilgan AR, Jernigan RL, Erman B. & Bahar, I. (1999) Protein Science 7, 2522. Compare experimental B-factors with theoretical B-factors theoretical B-factor experimental B-factor 50 40 30 20 10 0 0 50 100 150 residue number http://www.ccbb.pitt.edu/CCBBResearchDynHemRel.htm 200 250 300 Comparison of the slowest modes of T and R2 chain chain T 92-100 R2 35-40 b2 84-94 2 145-146 37-44 1 132-141 0 50 100 150 residue number 200 250 300 T R transitions in Hb Experimental T Experimental R2 Reference... Xu & Bahar, submitted. Computed (R2) Si = 3/2 <cos2Di> - 1/2 Order parameter Order parameters for CO-bound and unliganded Hb CO-Hb () deoxygenated ( ) 50 Order parameter 0 residue index 100 150 -1 deoxygenated Hb -1 CO-Hb For details on theory see... 0 Haliloglu & Bahar, Proteins 1999, 37, 654-667 30 60 90 residue index 120 150 Fluctuations of the nevirapine-bound (A) and unliganded (B) forms of RT (I) (A) (II) • Fluctuating conformations of the nevirapine-bound (A) and unliganded (B) forms of RT. The p66 subdomains are colored cyan (fingers), yellow (palm), red (thumb), green (connection) and pink (RNase H). (I) (B) (II) • See the difference in the mechanism of global fluctuations for the liganded and unliganded RTs. This difference is significant given that the sizes or distributions of fluctuations are unaffected by ligand binding http://www.ccbb.pitt.edu/CCBBResearchDomMot.htm Two hinge bending sites • (A) Two hinge-bending centers on RT forming minima in Figure 1: (I) near the NNRTI binding site, involving residues 107110 (cyan), 161-165 (green), 180-188 (red) and 219-231 (blue), and (II) near the p66 connection and RNase H interface, comprising residues 363-366 (cyan), 394-408 (green), 410-423 (loop, magenta), 424-429 (interdomain linker, red), and 504-512 (yellow). •(B) A closer view of region II, showing explicitly the side chains near the hinge site. Close tertiary contacts are indicated by the yellow dots. Topology-based models • • • • • • • • Near-native fluctuations • Ben-Avraham (1993) Tirion (1996) Bahar et al. (1997) Hinsen (1998) Sanejouand, Tama (2000) Wriggers, Brooks (2001) Ma (2002) • • • • • • • • Folding/unfolding processes (folding loss of configurational entropy) (springs acting on effective centroids, usually C atoms) Micheletti et al, PRL (1999) Cecconi et al. Proteins (2001) Go & Scheraga Macromolecules (1976) Galzitskaya & Finkelstein, PNAS (1999) Munoz et al. PNAS (1999) Alm & Baker, PNAS (1999) Klimov & Thirumalai, PNAS (2000) Clementi et al (Onuchic), JMB (2000) “Native topology determines forceinduced unfolding pathways” Protein folding kinetics examined by a Go-like model Koga, N. & Takada, S. J Mol. Biol. 2001, 313, 171-180 Topological and Energetic Factors: What determines the transition state ensemble, and folding intermediates? Simulations with Go-like potential Applied to CI2, SH3 (2-state folders) and barnase, RNase H and CheY (have intermediates) “ Topology plays a central role in determining folding mechanisms” Can we use such simplified approaches for estimating amyloidogenic intermediates? Clementi, C. Nyemeyer, H. & Onuchic, J. N. J Mol. Biol 2000, 298, 937.