Basics of Fluorescence
advertisement

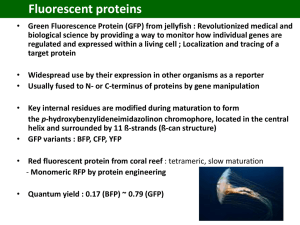
Today’s Announcements (In random order), students giving mid-term talks: Thuy Ngo Wylie Ahmed Charles Wilson Mohamed Ghoneim Xin Tang Claire Mathis Pengfei Yu Matthias Smith Anthony Hung Yu Ho Vishal Soni Joshua Glaser Eric Johnson Kiran Girdhar Email me your Powerpoint presentation to me at least 1 hr before class ! (Put your name as filename.) You may use your own computers but it may be harder. Today’s take-home lessons (i.e. what you should be able to answer at end of lecture) 1. 2. 3. 4. GFP can Fluoresce. Basics of labeling in vivo (GFP, FLASH, others…). Super-Accuracy (FIONA) Total Internal Reflection Green Fluorescent Protein (Nobel, 2009) Genetically encoded dye (fluorescent protein) (Motor) protein Kinesin – GFP fusion GFP Wong RM et al. PNAS, 2002 Genetically encoded perfect specificity Photo-active GFP G. H. Patterson et al., Science 297, 1873 -1877 (2002) Photoactivatable variant of GFP that, after intense irradiation with 413nanometer light, increases fluorescence 100 times when excited by 488nanometer light and remains stable for days under aerobic conditions Native= filled circle Photoactivated= Open squares Wild-type GFP T203H GFP: PAGFP GFP: How protein makes color Threonine (Thr or T)is an α-amino acid, HO2CCH(NH2)CH(OH)CH3. Its codons are ACU, ACA, ACC, and ACG. This essential amino acid is classified as polar. Tyrosine (abbreviated as Tyr or Y) is a nonessential amino acid with a polar side group. The word "tyrosine" is from the Greek tyros, meaning cheese, as it was first discovered in 1846 by German chemist Justus von Liebig in the protein casein from cheese. Glycine (Gly or G) , NH2CH2COOH, is the smallest of the 20 amino acids. Basics of Labeling In vivo (inside cell) Cell has a membrane, which is, in general, impermeant to dyes! Bi-Arsenic FLASH, Fluorescent Proteins, SNAP-tag, Halo-tag Bi-Arsenic FLASH, ReASH… Tsien, Science, 1998 Tsien, Science, 2002 Imaging (Single Molecules) with very good S/N (at the cost of seeing only a thin section very near the surface) Total Internal Reflection (TIR) Microscopy TIR- ( > c) Exponential decay dp=(l/4p)[n12sin2i) - n22]-1/2 For glass (n=1.5), water (n=1.33): TIR angle = >57° Penetration depth = dp = 58 nm With dp = 58 nm , can excite sample and not much background. Experimental Set-up for TIR (2 set-ups) Laser Sample Objective Sample Objective Dichroic Laser Filter Lens Filter CCD Detector CCD Detector Lens Wide-field, Prism-type, TIR Microscope Wide-field Objective-TIR Objective TIR: better S/N www.olympusmicro.com Fluorescence Imaging with One Nanometer Accuracy Very good accuracy: 1.5 nm, 1-500 msec Diffraction limited spot: Single Molecule Sensitivity Accuracy of Center = width/ S-N = 250 nm / √104 = 2.5 nm = ± 1.25nm Width of l/2 ≈ 250 nm Prism-type TIR 0.2 sec integration center 280 240 Photons 200 160 120 width 80 40 0 5 10 15 Y ax is 15 10 20 20 25 25 5 0 ta X Da Enough photons (signal to noise)…Center determined to ~1.3 nm Z-Data from Columns 1-21 Dye lasts 5-10x longer -- typically ~30 sec- 1 min. (up to 4 min) Start of high-accuracy single molecule microscopy Thompson, BJ, 2002; Yildiz, Science, 2003 How well can you localize? Depend on 3 things 1. # of Photons Detected (N) Prism-type TIR 0.2 sec integration center 280 2. Pixel size of Detector (a) 240 Photons 200 3. Noise (Background) of Detector (b) 160 120 width (includes background fluorescence and detector noise) 80 40 0 5 10 Y ax 15 is 15 20 20 25 25 10 ta X Da 5 0 = i s i2 a 2 12 8ps i4 b 2 2 2 N N a N Z-Data from Columns 1-21 derived by Thompson et al. (Biophys. J.). Class evaluation 1. What was the most interesting thing you learned in class today? 2. What are you confused about? 3. Related to today’s subject, what would you like to know more about? 4. Any helpful comments. Answer, and turn in at the end of class.