Claire_SfN_2013_PosterFINAL
advertisement
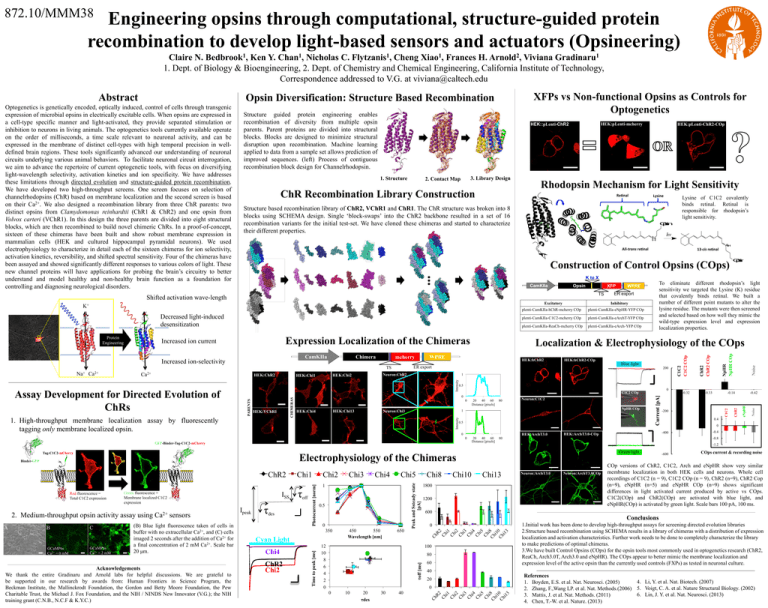
872.10/MMM38 Engineering opsins through computational, structure-guided protein recombination to develop light-based sensors and actuators (Opsineering) Claire N. Bedbrook1, Ken Y. Chan1, Nicholas C. Flytzanis1, Cheng Xiao1, Frances H. Arnold2, Viviana Gradinaru1 1. Dept. of Biology & Bioengineering, 2. Dept. of Chemistry and Chemical Engineering, California Institute of Technology, Correspondence addressed to V.G. at viviana@caltech.edu XFPs vs Non-functional Opsins as Controls for Optogenetics Opsin Diversification: Structure Based Recombination Structure guided protein engineering enables recombination of diversity from multiple opsin parents. Parent proteins are divided into structural blocks. Blocks are designed to minimize structural disruption upon recombination. Machine learning applied to data from a sample set allows prediction of improved sequences. (left) Process of contiguous recombination block design for Channelrhodopsin. HEK:pLenti-mcherry HEK::pLenti-ChR2 1. Structure 3. Library Design 2. Contact Map ChR Recombination Library Construction Rhodopsin Mechanism for Light Sensitivity Re nal O p s in hv N H All-trans re nal K to X CamKIIa Opsin XFP TS Decreased light-induced desensitization Expression Localization of the Chimeras 0.5 Neuron:Chi3 20 40 60 Distance [pixels] 80 NpHR COp 1 1. High-throughput membrane localization assay by fluorescently tagging only membrane localized opsin. 0.5 HEK:ArchT3.0 0 0 GFP-Binder-Tag-C1C2-mCherry Acknowledgements We thank the entire Gradinaru and Arnold labs for helpful discussions. We are grateful to be supported in our research by awards from: Human Frontiers in Science Program, the Beckman Institute, the Mallinckrodt Foundation, the Gordon and Betty Moore Foundation, the Pew Charitable Trust, the Michael J. Fox Foundation, and the NIH / NINDS New Innovator (V.G.); the NIH training grant (C.N.B., N.C.F & K.Y.C.) 0 -0.4 -0.8 Green light 0.5 0 Chi8 450 550 Wavelength [nm] 650 12 10 8 6 4 2 0 Chi10 Chi13 1800 1200 0 80 60 20 τdes 30 40 COp versions of ChR2, C1C2, Arch and eNpHR show very similar membrane localization in both HEK cells and neurons. Whole cell recordings of C1C2 (n = 9), C1C2 COp (n = 9), ChR2 (n=9), ChR2 Cop (n=9), eNpHR (n=5) and eNpHR COp (n=9) shows significant differences in light activated current produced by active vs COps. C1C2(COp) and ChR2(COp) are activated with blue light, and eNpHR(COp) is activated by green light. Scale bars 100 pA, 100 ms. 1.Initial work has been done to develop high-throughput assays for screening directed evolution libraries 2.Structure based recombination using SCHEMA results in a library of chimeras with a distribution of expression localization and activation characteristics. Further work needs to be done to completely characterize the library to make predictions of optimal chimeras. 3.We have built Control Opsins (COps) for the opsin tools most commonly used in optogenetics research (ChR2, ReaCh, Arch3.0T, Arch3.0 and eNpHR). The COps appear to better mimic the membrane localization and expression level of the active opsin than the currently used controls (FXPs) as tested in neuronal culture. 40 20 0 10 Neuron:ArchT3.0COp COps current & recording noise Conclusions 100 0 Neuron:ArchT3.0 -600 600 Ch R2 Ch i1 Ch i2 Ch i3 Ch i4 Ch i5 Ch i Ch 8 i1 Ch 0 i1 3 Cyan Light ChR2 Chi2 Chi5 1 350 Chi4 Chi4 Ch R2 Ch i1 Ch i2 Ch i3 Ch i4 Ch i5 Ch i Ch 8 i1 Ch 0 i1 3 GCaMP6s Ca2+ = 2 mM (B) Blue light fluorescence taken of cells in buffer with no extracellular Ca2+, and (C) cells imaged 2 seconds after the addition of Ca2+ for a final concentration of 2 mM Ca2+. Scale bar 20 μm. τdes Chi3 τoff [ms] GCaMP6s Ca2+ = 0 mM C sensors Ipeak τoff Chi2 Peak and Ssteady state [pA] 2. Medium-throughput opsin activity assay using ISS Time to peak [ms] Green fluorescence = Membrane localized C1C2 expression Chi1 Photocurrent [norm] ChR2 Ca2+ -0.42 0.4 80 Electrophysiology of the Chimeras Binder-GFP B 20 40 60 Distance [pixels] -200 -400 HEK:ArchT3.0-COp -0.14 -0.33 -1.2 Tag-C1C2-mCherry A Neuron:C1C2 -0.32 Noise 0 HEK:Chi13 0 C1C2 COp eNpHR Intensity 1 Noise 200 0 HEK:Chi4 HEK:hChR2-COp Blue light Intensity CHIMERAS PARENTS HEK:VChR1 plenti-CamKIIa-eArch-YFP COp HEK:hChR2 Neuron:ChR2 HEK:Chi2 HEK:Chi1 plenti-CamKIIa-ReaCh-mcherry COp ChR2 HEK:ChR2 plenti-CamKIIa-eArchT-YFP COp ER export TS Ca2+ plenti-CamKIIa-C1C2-mcherry COp Localization & Electrophysiology of the COps WPRE mcherry plenti-CamKIIa-eNpHR-YFP COp NpHR NpHR COp Chimera Inhibitory C1C2 CamKIIa ER export plenti-CamKIIa-hChR-mcherry COp O p s in To eliminate different rhodopsin’s light sensitivity we targeted the Lysine (K) residue that covalently binds retinal. We built a number of different point mutants to alter the lysine residue. The mutants were then screened and selected based on how well they mimic the wild-type expression level and expression localization properties. WPRE ChR2 ChR2 COp Increased ion current Increased ion-selectivity Red fluorescence = Total C1C2 expression 13-cis re nal Construction of Control Opsins (COps) Excitatory Assay Development for Directed Evolution of ChRs Lysine of C1C2 covalently binds retinal. Retinal is responsible for rhodopsin’s light sensitivity. N H K+ Na+ Ca2+ Lysine Structure based recombination library of ChR2, VChR1 and ChR1. The ChR structure was broken into 8 blocks using SCHEMA design. Single ‘block-swaps’ into the ChR2 backbone resulted in a set of 16 recombination variants for the initial test-set. We have cloned these chimeras and started to characterize their different properties. Shifted activation wave-length Protein Engineering HEK:pLenti-ChR2-COp C1C2 C1C2 COp Optogenetics is genetically encoded, optically induced, control of cells through transgenic expression of microbial opsins in electrically excitable cells. When opsins are expressed in a cell-type specific manner and light-activated, they provide separated stimulation or inhibition to neurons in living animals. The optogenetics tools currently available operate on the order of milliseconds, a time scale relevant to neuronal activity, and can be expressed in the membrane of distinct cell-types with high temporal precision in welldefined brain regions. These tools significantly advanced our understanding of neuronal circuits underlying various animal behaviors. To facilitate neuronal circuit interrogation, we aim to advance the repertoire of current optogenetic tools, with focus on diversifying light-wavelength selectivity, activation kinetics and ion specificity. We have addresses these limitations through directed evolution and structure-guided protein recombination. We have developed two high-throughput screens. One screen focuses on selection of channelrhodopsins (ChR) based on membrane localization and the second screen is based on their Ca2+. We also designed a recombination library from three ChR parents: two distinct opsins from Clamydomonas reinhardtii (ChR1 & ChR2) and one opsin from Volvox carteri (VChR1). In this design the three parents are divided into eight structural blocks, which are then recombined to build novel chimeric ChRs. In a proof-of-concept, sixteen of these chimeras have been built and show robust membrane expression in mammalian cells (HEK and cultured hippocampal pyramidal neurons). We used electrophysiology to characterize in detail each of the sixteen chimeras for ion selectivity, activation kinetics, reversibility, and shifted spectral sensitivity. Four of the chimeras have been assayed and showed significantly different responses to various colors of light. These new channel proteins will have applications for probing the brain’s circuitry to better understand and model healthy and non-healthy brain function as a foundation for controlling and diagnosing neurological disorders. Current [pA] Abstract References 4. Li, Y. et al. Nat. Biotech. (2007) 1. Boyden, E.S. et al. Nat. Neurosci. (2005) 2. Zhang, F.,Wang LP. et al. Nat. Methods.(2006) 5. Voigt, C. A. et al. Nature Structural Biology. (2002) 6. Lin, J. Y. et al. Nat. Neurosci. (2013) 3. Mattis, J. et al. Nat. Methods. (2011) 4. Chen, T.-W. et al. Nature. (2013)