wrky-for-balaji - MRC Laboratory of Molecular Biology
advertisement
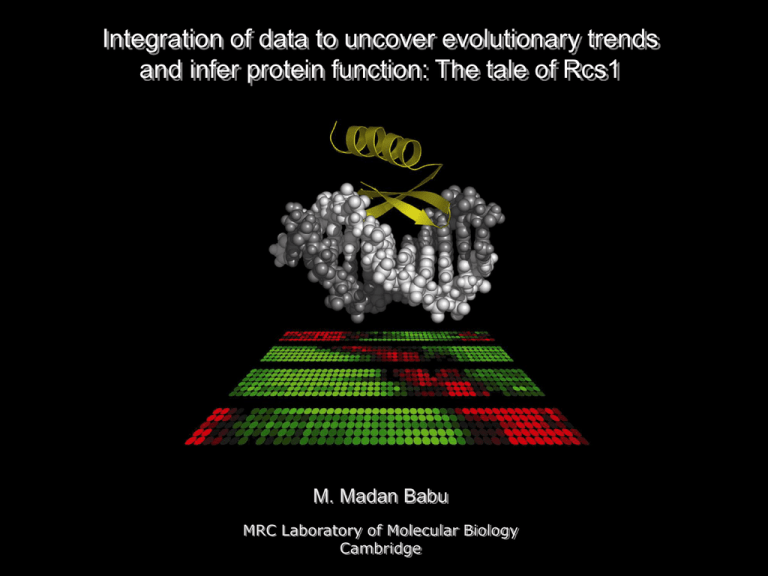
Integration of data to uncover evolutionary trends and infer protein function: The tale of Rcs1 M. Madan Babu MRC Laboratory of Molecular Biology Cambridge Overview of research Evolution of biological systems Evolution of networks within and across genomes Evolution of transcription factors Evolutionary of transcriptional networks Nuc. Acids. Res (2003) Nature Genetics (2004) J Mol Biol (2006a) Structure and function of biological systems Structure and dynamics of transcriptional networks J Mol Biol (2006b) J Mol Biol (2006c) Uncovering a distributed architecture in networks Methods to study network dynamics Nature (2004) Data integration, function prediction and classification Discovery of transcription factors in Plasmodium Discovery of novel DNA binding proteins C Evolution of a global regulatory hubs H H C Nuc. Acids. Res (2005) Cell Cycle (2006) Rcs1 – regulator of cell size 1 S. cerevisiae - wild type Size of mutant cells are twice that of the parental strain S. cerevisiae - Rcs1 mutant The critical size for budding in the mutant is similarly increased Rcs1 binds specific DNA sequences The following parameters that were used to define cell-size for the Rcs1 mutant were at least 2 Standard deviation (2 s) from the mean values of the wild-type Mother cell-size 874 760 Contour length of mother cell 108 100 Long axis length of mother cell 36 33 Short axis length of mother cell 30 27 Roundness of mother cell 1.29 1.20 Micrographs and data from SCMD Rcs1 is a global regulatory hub – Network analysis I P53 Tigger Dal82 Ime1 Tea Abf1 Tig AT-Hook Ace1 Rcs1 Gcr1 LisH HMG1 Mads Myb Apses Hsf Fkh bHLH Gata Homeo bZip C2H2-Zn C6-Fungal No. of members Rcs1p and Aft2p are global regulatory hubs with an as yet uncharacterized DNA binding domain Distribution of DNA binding domains in yeast transcription factors Transcriptional regulatory network in yeast Sub-network of Rcs1 and Aft2 Aft2p 123 41 Rcs1p 314 Number of target genes regulated How did the paralogous hubs that regulate distinct sets of genes evolve? Relationship to WRKY DNA binding domain – Sequence analysis I ... + . Non-redundant database Candida albicans (ascomycete) Yarrowia lipolytica (ascomycete) Ustilago maydis (basidiomycete) Cryptococcus sp (basidiomycetes) E. cuniculi (microsporidia) Giardia lamblia (diplomonad) Dictyostelium discoideum Entamoeba histolytica Lineage specific expansion in several fungi and is seen in lower eukaryotes WRKY domain (Arabidopsis) + FAR-1 type transposase (Medicago truncatula) Profiles + HMM of this region Non-redundant database Globular region maps to WRKY DNA-binding domain Confirmation of relationship to WRKY DBD – Sequence analysis II Rcs1 (S. cerevisiae) + WRKY DNA-binding Domain from Arabidopsis WRKY4 Gcm1 (Drosophila) Non-redundant database WRKY DNA-binding domain maps to the same globular region S1 S2 S3 S4 JPRED/PHD Multiple sequence alignment of all globular domains Sequence of secondary structure is similar to the WRKY DNA-binding domain and GCM1 protein seen in mouse Homologs of the conserved globular domain constitutes a novel family of the WRKY DNA-binding domain Characterization of the globular domain – structural analysis I Predicted SS of Rcs1 DBD S1 S2 S3 Predicted SS of Rcs1 DBD S4 S1 SS of WRKY4 S1 S2 S3 S2 S3 S4 SS of GCM1 S4 S1 S2 S3 S4 Template structure A. thaliana transcription factor (WRKY4:1wj2:NMR structure) Mus musculus Glial Cell Missing - 1 (GCM-1:1odh:X-ray structure) Both WRKY and GCM1 have similar network of stabilizing interactions Characterization of the globular domain – structural analysis II S1 S2 S3 S4 4 residues involved in metal co-ordination and 10 residues involved in key stabilizing hydrophobic interactions that determine the path of the backbone in the four strands of the GCM1-WRKY domain show a strong pattern of conservation. Core fold of the Rcs1 DBD will be similar to the WRKY-GCM1 domain and may bind DNA in a similar way Classification of WRKY-GCM1 superfamily – Cladistic analysis I S1 S2 S3 S4 C H Zn2+ + H C S1 S2 S3 S4 Template structure Classical WRKY (C) C Insert containing version (I) HxC containing version (HxC) C H C Zn2+ Zn2+ C C C S2 S3 S4 WRKY motif in S1 Short loop between S2 & S3 C WRKY4 S1 C H W S2 S3 H C I S1 S2 H S3 S1 S4 S2 Rcs1 HxC instead of HxH N-terminal helix Short insert between S2 & S3 HxC H C S3 S4 Conserved W in S2 Sequence features F H Zn2+ C W S4 N-terminal helix Conserved W in S4 Large insert between S2 & S3 Far1 H Zn2+ C S1 GCM domain (G) H Zn2+ H FLYWCH domain (F) Mdg S1 S2 S3 S4 Insertion of Zn ribbon between S2 and S3 G Gcm1 Domain context for the different families – network analysis I HxC containing version (HxC) C H Zn2+ C H C Zn2+ H Zn2+ H C W S2 S3 OUT protease MULE Tpase Zn knuckle SMBD Stand alone I Stand alone Tandem e.g. WRKY4 I Tandem C I Zn cluster C e.g. Rcs1 S2 S3 S1 S4 S2 S3 S4 S1 S2 S3 e.g. At2g23500 HxC F G HxC F G e.g. 101.t00020 H H C S4 e.g. Far1 C S1 BED finger S1 Zn2+ Mobile element S4 Stand alone S3 MULE Tpase S2 H C W C S1 H H Zn2+ C C GCM domain (G) e.g. Mod (mdg) Stand alone C FLYWCH domain (F) POZ C Insert containing version (I) Stand alone Classical WRKY (C) e.g. Gcm1 S4 C I TF only TF only TF + TP Phyletic distribution – Comparative genome analysis I HxC F Transcription factor G Transposase Human Fly Higher Eukaryotes GCM1 and FLYWCH versions evolved from an insert containing version that is a transposase Worm Fungi Fungi Entamoeba Lower eukaryotes Slim mould Plants Plants HxC and Insert containing versions are seen as both transcription factors and as transposases Classical version of the WRKY evolved from an insert containing version that is a transposase -explain that there has been multiple transitions from transposase to TFs in the fungal genomes -explain how this could have happened by showing the snapshot of the breakup of selfish elements into two distinct products -explain that the transposase can itself regulate the gene expression of itself Outline of the presentation Rcs1 and aft2 have a distinct version of the WRKY type DNA binding domain Sensitive sequence search reveals that Oryza sativa (monocot) Arabidopsis thaliana (dicot) Medicago truncatula (dicot) Nicotiana tabacum (dicot) Structural equivalences of WRKY-GCM1 domain proteins with Bed and Zn finger WRKY (1wj2) GCM-type WRKY (1odh) Zn C Zn2+ S2 C C H Zn2+ H C S1 H C H S1 S4 S2 C C H C H Zn2+ Zn2+ C S3 Classical Zn-finger (1m36) C C C Bed-finger (2ct5) H H S3 S4 S1 S2 S3 S4 S1 S2 H1 Why Rcs1? While systematically analyzing the genes which gave rise to abnormal cell size, We and the other noted that mutants of Rcs1 give abnormal cell shape. It was known to be an important transcription factor involved in cell size regulation – explain showing graphs and images Independently, during the analysis of the TNET in yeast We looked at the hubs and the DNA binding domains That were present in them. Interestingly, there were two Hubs that did not have any known DNA binding domain Identified in them, but the region which mediates DNA was known – explain showing the family relationship Of the hubs -only two members, and both are hubs -how and when did they evolve? Standard search procedures using Pfam and other databases did not provide any clue about the domain. So we set out to characterize the DNA binding region from Rcs1p and its paralog Aft2p using sensitive sequence search and other computational methods. -show output from Pfam hits WRKY DNA binding domain – Structure analysis I Structural aspects of the DNA binding domain Explain the residues involved in metal chelating -DNA contacting surface -Inserts in the loops -Stabilizing contacts involved WRKY DNA binding domain – Structure analysis II Structure comparisons identify several other Known transcription factors including the GCM protein in eukaryotes -Explain the insert of a zinc ribbon in the loop In fact sequence comparison without the insert can pick these WRKY proteins Classification of WRKY domains – Cladistic analysis I Multiple starting points identified all homologs in the different species This allowed us to classify the sequences into different families Each with a specific feature suggesting common evolutionary relationship Based on shared and derived features of the domains - List the 5 families and point to features involved using a structure template Phylogenetic distribution and domain architecture for the different families - I Phyletic profiles of the different domains points to the possibility that these transcription factors could have evolved from transposases With at least two distinct recruitment into transcription factors. -In plants in one case -In the base of the fungal genomes in the other case Phylogenetic distribution and domain architecture for the different families - II Comparative genomics using the fungal genomes provides the clue for the evolution of these TFs -explain that there has been multiple transitions from transposase to TFs in the fungal genomes -explain how this could have happened by showing the snapshot of the breakup of selfish elements into two distinct products -explain that the transposase can itself regulate the gene expression of itself Comparative genomics using the fungal genomes provides the clue for the evolution of these TFs -extensive recruitment of the transposase in the different fungal lineages -multiple jumps within the fungal lineage -very recent duplication event in the order Saccharomycetales suggest hubs could Evolve rapidly -Candida rbf1 and other TFs independently duplicated and evolved as global regulators Analysis of the gene expression data in plants Since it happened in fungal genomes, we ask how does this behave in the plants. -show the gene expression patterns for the different subfamilies. We see two trends one where divergence has primarily occurred in the expression changes rather than in the protein sequence, and the other in which proteins with the same expression pattern have different binding site residues. -spatio-temporal changes in gene expression -It is experimentally well known that the FLYWCH and the GCM proteins are developmentally important regulatory proteins. So in three lineages there has been recruitment of the transposase into becoming a developmentally important global regulator. Analysis of the gene expression data in plants There are interesting traces of gene expression pattern when we see for the different WRKY containing proteins. TPases are expressed in the root and in the pollen enhancing the possibility of rapidly expanding themselves during evolution. Acknowledgements Aravind group L Aravind S Balaji Lakshminarayan Iyer MtrDRAFT_AC146590g49v2_Mtru_92891293 * I C hGCMa_Hsap_1769820 * C * NtEIG-D48_Ntab_10798760 mod(mdg4)_Dmel_24648712 F 1- 5 CG13845_Dmel_24649011 Homo sapiens I Drosophila melanogaster I I * * I YALI0C00781g_Ylip_50547661 I C26E6.2_Cele_32565510 F I HxC I Ci-ZF-1_Cint_93003122 F 1- 5 KIAA1552_Hsap_10047169 LOC_Os11g31760_Osat_77551147 C20orf164_Hsap_13929452 C CHGG_00311_Cglo_88184608 I LOC411361_Amel_66547010 F HxC CHGG_08318_CGLO_88179597 I I T24C4.2_Cele_17555262 * UM03656.1_Umay_71019145 I Caenorhabditis elegans AN6124.2_ANID_67539908 FAR1_Atha_18414374 I AT4g19990_Atha_7268794 C F G WRKY41_Osat_46394336 At2g23500_Atha_3242713 * I F54C4.3_Cele_3790719 * TTR1_Atha_30694675 I gcm_Dmel_17137116 MtrDRAFT_AC126008g21v1_Mtru_92876827 C YALI0A02266g_Ylip_50543034 T24C4.7_Cele_17555272 G * I I HxC Fungi Plants HxC C C Animals WRKY58_Atha_22330782 At2g34830_Atha_27754312 I mutA_Ylip_49523824 I Afu2g08220_Afum_71000950 AFT2_Scer_6325054 Encephalitozoon cuniculi ECU05_0180_Ecun_19173554 Ciliates HxC Apicomplexa I Giardia lamblia I 101.t00020_Ehis_67474280 GLP_9_36401_35940_Glam_71071693) Entamoeba histolytica C C C Classical WRKY GCM-type G WRKY C I F Insert-containing HxC WRKY FLYWCH-type WRKY HxC-type WRKY MULE transposase C Dictyostelium dd_03024_Ddis_28829829 discoideum GLP_79_64671_67418_Glam_71077115) Plant specific Zn-cluster Zinc knuckle BED finger SWIM domain Plant-specific mobile domain PHD finger C2H2 finger LRR STAND ATPase Isochoris matase Plant specific N-all-beta TIR domain AT-hook OTU POZ Expression profiles of WRKY-GCM1 domain proteins in Arabidopsis Gene expression profiles for the light exposure conditions in Arabidopsis thaliana + WRKY proteins show light specific expression 15 Far1-type proteins 15 Far1-type proteins 5 WRKY domain Proteins with TIR/LRR 5 WRKY domain Proteins with TIR/LRR 60 WRKY domain containing proteins b + 40 HxC type WRKY domain proteins 40 HxC type WRKY domain proteins WRKY proteins show tissue specific expression Gene expression profiles for the developmental stages in Arabidopsis thaliana 60 WRKY domain containing proteins a ot tem S Ro f a Le ex er al Ap low or F Fl ans g or Se s ed ess ous rkn tinu a D Con ght li lse Pu ht lig Relationship between Rcs1p and Aft2p homologs Multiple independent evolution of TFs from Transposons UM03656.1 Umay 71019145 CAGL0H03487G CGLA 49526254 CAGL0G09042G CGLA 49526062 CaO19.2272 Calb 68482460 DEHA0F25124g Dhan 50425555 KLLA0D03256g Klac 50306475 AFL087C AGOS 44984319 ORFP Sklu Contig1830.2 kluyveri Kwal 24045 waltii ORFP Scas Contig720.21 castelli ORFP Skud Contig2057.12 kudriavzeii ORFP 7853 mikatae * ORFP 8601 paradoxus RCS1 SCER 51830313 ORFP Scas Contig690.14 castelli Rcs1 Aft2p cluster ORFP Skud Contig1659.3 kudriavzeii Animals Rbf1 cluster ORFP 21513 mikatae Plants * ORFP 22109 paradoxus Entamoeba AFT2 SCER 6325054 Fungi AAL026Wp Agos 44980144 UM03656.1 Umay 71019145 CHGG 06963 CGLO 88178242 CHGG 06785 CGLO 88182698 CHGG 09478 CGLO 88177996 CHGG 00175 CGLO 88184472 CHGG 10902 CGLO 88175616 FG05699.1 Gzea 46122643 NCU06551.1 Ncra 85106835 NCU05145.1 Ncra 85081010 YALI0F07128g Ylip 50555399 MG05295.4 Mgri 39939890 FG04147.1 Gzea 46116610 NCU07855.1 Ncra 85109845 MG06795.4 Mgri 39977821 NCU08168.1 Ncra 85093270 CHGG 09951 CGLO 88176079 CHGG 08318 CGLO 88179597 NCU04492.1 Ncra 32406464 FG09606.1 Gzea 46136181 NCU06975.1 Ncra 85108658 CHGG 05063 CGLO 88180976 HOP78 FOXY 30421204 CHGG 00311 CGLO 88184608 CIMG 00825 CIMM 90305840 AN6124.2 Anid 67539908 ISOCHOR AFUM 71001046 CNC00740 CNEO 57225606 CNBH2400 Cneo 50256416 AN0859.2 ANID 67517161 YALI0A16269g Ylip 50545173 CaO19 12424 Calb 68467239 DEHA0E17127g Dhan 50422877 RBF1P CALB 2498834 DEHA0A05258g Dhan 50405817 CaO19.2272 Calb 68482460 DEHA0F25124g Dhan 50425555 CAGL0H03487G CGLA 49526254 AFL087C AGOS 44984319 KLLA0D03256g Klac 50306475 CAGL0G09042G CGLA 49526062 RCS1 SCER 51830313 AFT2 SCER 6325054 YALI0A05313g Ylip 50543230 YALI0A02266g Ylip 50543034 Mutyl Ylip 50545163 YALI0C17193g.c Ylip 50548927 Mutyl.c Ylip 50545161 YALI0C00781g.d Ylip 50547661 YALI0C00781g.a Ylip 50547661 YALI0C00781g.b Ylip 50547661 YALI0C00781g.c Ylip 50547661 YALI0C17193g.a Ylip 50548927 Mutyl.a Ylip 50545161 YALI0D22506g Ylip 50551361 Mutyl.b Ylip 50545161 YALI0C17193g.b Ylip 50548927 MG07557.4 Mgri 39972511 MG09992.4 Mgri 39965911 101.T00020 EHIS 67474280 4.T00052 EHIS 67483840 FAR1 ATHA 18414374 AT2G27110 ATHA 18401324 AT2G43280 ATHA 30689328 AT4G38180 ATHA 15233732 AT3G59470 ATHA 18411179 AT5G28530 ATHA 22327146 AT1G52520 ATHA 15219020 AT1G80010 ATHA 15220043 C20ORF164 HSAP 13929452 LOC428161 GGAL 50759053 T24C4.2 CELE 17555262 SJCHGC04823 SJAP 56758936 6330408A02RIK MMUS 50053999 LOC374920 HSAP 27694337 Transcriptional network involving Aft2p and Rcs1p Aft2p Aft2p Rcs1p Rcs1p 123 41 314 Number of target genes regulated Conclusion Integration of different types of experimental data allowed us to Identify the DNA binding domain in Rcs1 Sequence Structure Expression Interaction