run
advertisement
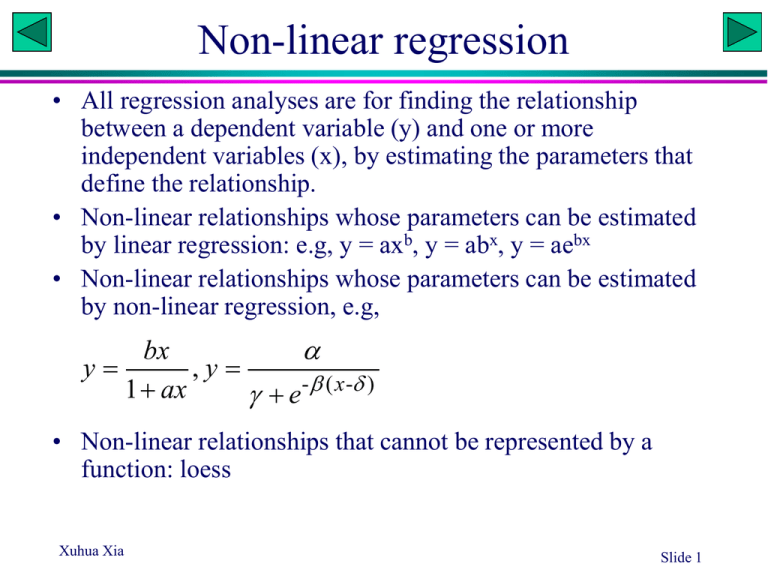
Non-linear regression • All regression analyses are for finding the relationship between a dependent variable (y) and one or more independent variables (x), by estimating the parameters that define the relationship. • Non-linear relationships whose parameters can be estimated by linear regression: e.g, y = axb, y = abx, y = aebx • Non-linear relationships whose parameters can be estimated by non-linear regression, e.g, bx y ,y 1 ax e- ( x - ) • Non-linear relationships that cannot be represented by a function: loess Xuhua Xia Slide 1 Growth curve of E. coli • A researcher wishes to estimate the growth curve of E. coli. He put a very small number of E. coli cells into a large flask with rich growth medium, and take samples every half an hour to estimate the density (n/L). • 14 data points over 7 hours were obtained. • What is the instantaneous rate of growth (r). What is the initial density (N0)? • As the flask is very large, he assumed that the growth should be exponential, i.e., y = a·ebx (Which parameter correspond to r and which to N0?) Xuhua Xia Slide 3 SAS Program /* Fictitious data */ data Ecoli; Input Density @@; Time = _N_; lnD = log(Density); datalines; 20.023 39.833 80.571 161.102 317.923 635.672 1284.544 2569.430 5082.654 10220.777 20673.873 40591.439 81374.642 163963.873 ; proc reg; This statement is necessary, otherwise Density var Density lnD Time; will be an unknown variable for the reg model lnD = Time; procedure and the plot statement will fail. plot Density*Time/ symbol='.'; plot lnD*Time/ symbol='.'; Run, and ask students to compute run; parameters a and b from regression output. What is the initial density? Xuhua Xia Slide 4 Body weight of wild elephant • A researcher wishes to estimate the body weight of wild elephants. • He measured the body weight of 13 captured elephants of different sizes as well as a number of predictor variables, such as leg length, trunk length, etc. Through stepwise regression, he found that the inter-leg distance (shown in figiure) is the best predictor of body weight. • He learned from his former biology professor that the allometric law governing the body weight (W) and the length of a body part (L) states that W = aLb • Use linear regression to find parameters a and b. Xuhua Xia Slide 5 SAS Program /*Fitcitious data */ data Elephant; Input L W @@; lnL = log(L); lnW = log(W); datalines; 0.3 1.657 0.4 2.500 0.5 4.680 0.6 7.075 0.7 10.070 0.8 11.988 0.9 14.836 1 18.318 1.1 23.496 1.2 27.897 1.3 36.796 1.4 44.611 1.5 50.183 ; proc reg; Run and ask students to var L W lnL lnW; calculate the parameters a model lnW = lnL; and b. plot W*L/ symbol='.'; plot lnW*lnL/ symbol='.'; run; Xuhua Xia Slide 6 DNA and protein gel electrophoresis • How to estimate the molecular mass of a protein? – A ladder: proteins with known molecular mass – Deriving a calibration curve relating molecular mass (M) to migration distance (D): D = F(M) – Measure D and obtain M • The calibration curve is obtained by fitting a regression model Xuhua Xia Slide 7 Protein molecular mass • The equation appears to describe the relationship between D and M quite well. This relationship is better than some published relationships, e.g., D = a – b ln(M) • The data are my measurement of D and M for a subset of secreted proteins from the gastric pathogen Helicobacter pylori (Bumann et al., 2002). • Write a SAS program to use the data to find parameters a and b D ae bM Mass D 5 14.5 10 12.6 20 9.4 30 7.1 40 5.3 50 3.9 60 3.05 70 2.3 80 1.75 Bumann, D., Aksu, S., Wendland, M., Janek, K., Zimny-Arndt, U., Sabarth, N., Meyer, T.F., and Jungblut, P.R., 2002, Proteome analysis of secreted proteins of the gastric pathogen Helicobacter pylori. Infect. Immun. 70: 3396-3403. Xuhua Xia Slide 8 Area and Radius What is the functional relationship between the area and the radius? Xuhua Xia Toxicity study: pesticide 100 90 Percentage killed 80 70 60 50 40 30 20 10 0 25 30 35 40 45 50 55 60 65 70 Dosage What transformation to use? Xuhua Xia Slide 10 Probit and probit transformation • Probit has two names/definitions, both associated with standard normal distribution: – the inverse cumulative distribution function (CDF) – quantile function 0.9 0.8 0.7 0.6 CDF • CDF is denoted (z), which is a continuous, monotone increasing sigmoid function in the range of (0,1), e.g., (z) = p (-1.96) = 0.025 = 1 - (1.96) • The probit function gives the 'inverse' computation, formally denoted -1(p), i.e., probit(p) = -1(p) probit(0.025) = -1.96 = -probit(0.975) • [probit(p)] = p, and probit[(z)] = z. 1 0.5 0.4 0.3 0.2 0.1 0 -2.5 -1.5 -0.5 0.5 1.5 z Xuhua Xia Slide 11 SAS program Data Pesticide; Input Dosage Percent @@; Why NuProbit = probit(Percent/100); Cards; 27 0.90 28 1.39 31 2.40 31 2.49 35 6.42 36 7.78 37 9.16 38 10.21 38 11.71 40 16.24 41 16.90 43 22.94 44 27.35 44 27.45 44 28.14 45 28.97 45 29.96 45 30.50 46 34.30 46 35.39 46 35.65 47 37.55 47 38.46 48 40.97 49 44.37 49 45.71 49 46.66 49 47.38 50 49.86 50 52.26 51 55.12 51 56.12 52 57.68 52 59.99 52 60.30 53 60.51 53 61.82 53 62.00 53 62.92 54 66.06 54 67.14 55 70.58 55 71.57 56 74.11 56 74.12 57 76.77 57 77.01 58 78.56 58 79.01 59 83.53 60 83.96 60 84.40 61 87.95 62 88.74 63 91.13 64 92.64 64 92.67 66 95.49 68 97.00 69 97.15 ; Proc reg; Model Percent = Dosage / R CLM alpha = 0.01 CLI; Plot Percent*Dosage / symbol = '.'; Model NuProbit = Dosage / R CLM alpha = 0.01 CLI; Plot NuProbit*Dosage / symbol = '.'; run; Xuhua Xia divide Percent by 100? Run and explain CLM: CL of mean CLI: CL of individual observation Graphic contrast between the original and the transformed DV Slide 12 Non-linear regression • In rapidly replicating unicellular eukaryotes such as the yeast, highly expressed intron-containing genes requires more efficient splicing sites than lowly expressed genes. • Natural selection will operate on the mutations at the slicing sites to optimize splicing efficiency. • Designate splicing efficiency as SE and gene expression as GE. • Certain biochemical reasoning suggests that SE and GE will follow the following relationships: GE GE 2 if GE GE0 E[ SE | GE ] if GE GE0 c Xuhua Xia GE SE 1 0.46 2 0.47 3 0.57 4 0.61 5 0.62 6 0.68 7 0.69 8 0.78 9 0.7 10 0.74 11 0.77 12 0.78 13 0.74 13 0.8 15 0.8 16 0.78 Slide 14 Guess initial values GE GE 2 if GE GE0 E[ SE | GE ] c if GE GE 0 0.85 When GE=0 then SE = , so 0.4 0.8 When GE increases from 2 to 8, SE increases from 0.47 to 0.75, so (0.75-0.47)/(8-2) 0.047 0.75 0.7 With 0.4 and 0.047, then SE for GE = 12 should be 0.4+0.04712 = 0.96, but the actual SE is only about 0.77. This must be due to the quadratic term GE2, i.e., SE 0.65 0.6 0.55 0.5 (0.77 - 0.96) = 122, so 0.45 - 0.002 0.4 0 2 4 6 8 10 12 14 16 GE Xuhua Xia Slide 15 A few more twists GE GE 2 if GE GE0 E[ SE | GE ] if GE GE0 c The continuity condition requires that E[SE | GE0 ] GE0 GE02 The smoothness condition requires that E[ SE | GE0 ] 2 GE0 0 GE0 The two conditions implies that GE0 c 2 GE0 GE02 Xuhua Xia 2 4 Slide 16 SAS program /* Fictitious data */ data Intron; input SE GE @@; datalines; .46 1 .47 2 .57 3 .61 4 .62 5 .68 6 .69 7 .78 8 .70 9 .74 10 .77 11 .78 12 .74 13 .80 13 .80 15 .78 16 ; title 'Quadratic Model with Plateau'; proc nlin data=Intron; Initial guestimates parms alpha=.4 beta=.047 gamma=-.002; GE0 = -.5*beta / gamma; if (GE < GE0) then Y = alpha + beta*GE + gamma*GE*GE; else Y = alpha + beta*GE0 + gamma*GE0*GE0; A conditional statement model SE = Y; needed to model the two if _obs_ =1 and _iter_ =. then do; segments plateau =alpha + beta*GE0 + gamma*GE0*GE0; put / GE0= plateau= ; Write out GE0 and plateau (which could end; be computed from the estimated , , and output out=b predicted=SEp; . However, the output of these parameters run; Xuhua Xia computes predicted values for plotting and saves them to data set b contain rounding errors. Run and explain Slide 17 Plot proc sgplot data=b noautolegend; yaxis label='Observed or Predicted'; refline 0.7775 / axis=y label="Plateau" labelpos=min; refline 12.7476 / axis=x label="GE0" labelpos=min; scatter y=SE x=GE; series y=SEp x=GE; run; Xuhua Xia Slide 18 Fitting another function /* Fictitious data */ data Intron; input SE GE @@; datalines; .46 1 .47 2 .57 3 .61 4 .62 5 .68 6 .69 7 .78 8 .70 9 .74 10 .77 11 .78 12 .74 13 .80 13 .80 15 .78 16 ; title 'Quadratic Model with Plateau'; proc nlin data=Intron; parms alpha=.4 beta=.8 gamma=1; model SE = (alpha+beta*GE)/(1+gamma*GE); output out=b predicted=SEp; SE run; proc sgplot data=b noautolegend; yaxis label='Observed or Predicted'; scatter y=SE x=GE; series y=SEp x=GE; run; GE 1 GE Run and explain the output Xuhua Xia Slide 19 Plot output Xuhua Xia Slide 20 Robust regression • LOWESS: robust local regression between Y and X, with linear fitting • LOESS: robust local regression between Y and one or more Xs, with linear or quadratic fitting • Used with relations that cannot be expressed in functional forms • SAS: proc loess • Data: – Data set 1: monthly averaged atmospheric pressure differences between Easter Island and Darwin, Australia for a period of 168 months (NIST, 1998), suspected to exhibit 12-month (annual), 42month (El Nino), and 25-month (Southern Oscillation) cycles (From Robert Cohen of SAS Institute) – Data set 2: Two-channel microarray data. Background correction and two-channel loess normalization (from Wudu). Xuhua Xia Slide 21 SmoothingParameter=0.2 Pressure 18 17 16 15 14 13 12 11 10 9 8 7 6 5 4 3 2 1 0 0 10 20 30 40 50 60 70 80 90 Month 100 110 120 130 140 150 160 170 SmoothingParameter=0.05 Pressure 18 17 16 15 14 13 12 11 10 9 8 7 6 5 4 3 2 1 0 0 10 20 30 40 50 60 70 80 90 Month 100 110 120 130 140 150 160 170 Step 1: characterize the most obvious trend. data ENSO; input Pressure @@; Month=_N_; datalines; 12.9 11.3 10.6 11.2 10.9 7.5 7.7 11.7 12.9 14.3 10.9 13.7 17.1 14.0 15.3 8.5 5.7 5.5 7.6 8.6 7.3 7.6 12.7 11.0 12.7 12.9 13.0 10.9 10.4 10.2 8.0 10.9 13.6 10.5 9.2 12.4 12.7 13.3 10.1 7.8 4.8 3.0 2.5 6.3 9.7 11.6 8.6 12.4 10.5 13.3 10.4 8.1 3.7 10.7 5.1 10.4 10.9 11.7 11.4 13.7 14.1 14.0 12.5 6.3 9.6 11.7 5.0 10.8 12.7 10.8 11.8 12.6 15.7 12.6 14.8 7.8 7.1 11.2 8.1 6.4 5.2 12.0 10.2 12.7 10.2 14.7 12.2 7.1 5.7 6.7 3.9 8.5 8.3 10.8 16.7 12.6 12.5 12.5 9.8 7.2 4.1 10.6 10.1 10.1 11.9 13.6 16.3 17.6 15.5 16.0 15.2 11.2 14.3 14.5 8.5 12.0 12.7 11.3 14.5 15.1 10.4 11.5 13.4 7.5 0.6 0.3 5.5 5.0 4.6 8.2 9.9 9.2 12.5 10.9 9.9 8.9 7.6 9.5 8.4 10.7 13.6 13.7 13.7 16.5 16.8 17.1 15.4 9.5 6.1 10.1 9.3 5.3 11.2 16.6 15.6 12.0 11.5 8.6 13.8 8.7 8.6 8.6 8.7 12.8 13.2 14.0 13.4 14.8 ; ods output OutputStatistics=ENSOstats FitSummary=ENSOsummary; proc loess data=ENSO; The output data ENSOstats contains variables: model Pressure=Month / CLM SoothingParameter, Month, DepVar, Pred, etc. smooth = 0.02 to 0.2 by 0.01 dfmethod=exact; run; symbol1 c=black i=join value=dot; Three criteria for choosing the smooth parameter: symbol2 c=black i=join value=none; GCV: generalized cross-validation (Craven and Wahba proc gplot data=ENSOstats; 1979) by SmoothingParameter; plot (DepVar Pred)*Month/overlay; AIC: Akaike information criterion (Akaike 1973) run; AICC1: bias-corrected AIC (JUrvich and Simonoff 1988) Summary output: kd Tree, linear max(1, n*s/5) N. Fitting Points kd Tree Bucket Size Deg. Local Polynom. Smoothing Param. Neighbor Points RSS Trace[L] GCV AICC AICC1 Delta1 Delta2 Equivalent N. Param. Lookup DF Residual SE 168 168 168 168 105 105 1 1 1 1 2 2 1 1 1 1 1 1 0.02 0.03 0.04 0.05 0.06 0.07 3.0 5.0 6.0 8.0 10.0 11.0 0.0 293.1 468.8 603.6 737.2 736.7 168.0 72.3 49.1 37.1 29.6 29.4 . 0.0 0.0 0.0 0.0 0.0 . 3.1 2.9 2.9 2.9 2.9 . 567.8 496.1 487.4 495.1 494.5 0.0 82.3 110.1 124.4 132.6 132.8 0.0 78.6 108.3 123.5 130.0 130.2 168.0 58.9 40.3 30.6 23.8 23.7 . 86.1 111.8 125.3 135.2 135.4 . 1.9 2.1 2.2 2.4 2.4 105 65 65 2 3 3 1 1 1 0.08 0.09 0.1 13.0 15.0 16.0 868.5 1057.6 1196.5 24.7 21.1 18.8 0.0 0.0 0.1 3.0 3.1 3.2 507.2 530.0 544.1 138.6 142.6 145.6 136.9 140.3 143.8 20.1 16.9 15.1 140.3 144.9 147.4 2.5 2.7 2.9 int(n*s), where n is ‘N. Fitting Points’ and s is ‘Smotthing Param.’ Xuhua Xia Slide 25 More on Output 1 Trace( I L)T ( I L) 2 Trace ( I L) ( I L) T 2 12 / 2 Lookup Degrees of Freedom Residual SE= RSS/1 RSS AIC n ln 2p n RSS AICC1 n ln (greater penalty than 2 p) n Xuhua Xia Slide 26 SmoothingParameter=0.05 Pressure 18 17 16 15 14 13 12 11 10 9 8 7 6 5 4 3 2 1 0 0 10 20 30 40 50 60 70 80 90 Month 100 110 120 130 140 150 160 170 proc loess data=ENSO; model Pressure=Month / smooth = 0.01 to 0.2 by 0.01 dfmethod=exact degree = 2; run; SmoothingParameter=0.05 Pressure 18 17 16 15 14 13 12 11 10 9 8 7 6 5 4 3 2 1 0 0 10 20 30 40 50 60 70 80 90 Month 100 110 120 130 140 150 160 170 99% Confidence limits ods output OutputStatistics=ENSOstats FitSummary=ENSOsummary; proc loess data=ENSO; model Pressure=Month / r CLM smooth = 0.05 alpha=0.01 dfmethod=exact; run; symbol1 c=black i=none value=dot; symbol2 c=black i=join value=none; symbol3 c=red i=join value=none; symbol4 c=red i=join value=none; proc gplot data=ENSOstats; format DepVar 4.0; plot (DepVar Pred UpperCl LowerCL)*Month/ overlay hminor = 0 vminor = 0 vaxis = axis1 frame; run; Xuhua Xia Slide 29 99% confidence limits 20 Pressure 16 12 8 4 0 0 10 20 30 40 50 60 70 80 90 Month 100 110 120 130 140 150 160 170 Step 2: Characterize other trends /* modify data set ENSO to filter out the 12-month cycle */ data ENSO(drop=pi); set ENSO; pi = 4 * atan (1); /* or pi = 3.1415926 */ cos1 = cos(2*pi*Month/12); sin1 = sin(2*pi*Month/12); proc reg data=ENSO; model Pressure = cos1 sin1; r: residual, i.e., what is left after the output out=ENSO1 r=FilteredPressure; monthly cycle has been removed. run; proc print data=ENSO1; var Month Pressure FilteredPressure; run; ods output OutputStatistics=ENSO1stats; proc loess data=ENSO1; model FilteredPressure=Month/ smooth = 0.12; run; proc gplot data=ENSO1stats; plot (DepVar Pred)*Month/overlay; run; Xuhua Xia Slide 31 Cos1 and Sin1 1 0.8 0.6 0.4 Cos1, Sin1 0.2 Cos1 0 0 20 40 60 80 100 120 140 160 180 Sin1 -0.2 -0.4 -0.6 -0.8 -1 Month Xuhua Xia Slide 32 Filtered Pressure 20 15 Pressure 10 Pressure 5 FilteredP 0 -1 -0.8 -0.6 -0.4 -0.2 0 0.2 0.4 0.6 0.8 1 -5 -10 Cos(2*Pi*Month/12) Xuhua Xia Slide 33 ~42-month cycle 7 6 5 4 3 2 Residual 1 0 -1 -2 -3 -4 -5 -6 -7 -8 Xuhua Xia 0 10 20 30 40 50 60 70 80 90 Month 100 110 120 130 140 150 160 170 Slide 34 Any other trend? /* modify data set ENSO to filter out the 12-month cycle */ data ENSO(drop=pi); set ENSO; pi = 4 * atan (1); cos1 = cos(2*pi*Month/12); sin1 = sin(2*pi*Month/12); cos2 = cos(2*pi*Month/42); sin2 = sin(2*pi*Month/42); proc reg data=ENSO; model Pressure = cos1 sin1 cos2 sin2; output out=ENSO1 r=FilteredPressure; run; proc print data=ENSO1; var Month Pressure FilteredPressure; run; ods output OutputStatistics=ENSO1stats; proc loess data=ENSO1; model FilteredPressure=Month/ smooth = 0.12; run; proc gplot data=ENSO1stats; plot (DepVar Pred)*Month/overlay; run; Xuhua Xia Slide 35 ~25-month cycle 6 5 4 3 2 Residual 1 0 -1 -2 -3 -4 -5 -6 -7 0 Xuhua Xia 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160 170 Month Slide 36 Microarray • Diagnosis • Signature genes • Gene regulation pathway Avian coronavirus Xuhua Xia Bovine coronavirus Slide 37 Spotted cDNA Microarray cDNA “B” Cy3 labeled cDNA “A” Cy5 labeled + Laser 1 Scanning Hybridization DNA molecules are immobilized by high-speed robots on a solid surface such as glass. + Analysis Xuhua Xia Laser 2 Image Capture Slide 38 Microarray Data Number 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 Array Row Array Column Row 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 Xuhua Xia Column 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 2 2 2 2 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 1 2 3 4 Name X Location Y Location ch1 Intensity ch1 Background 61m12 Vacuolar ATP 2110synthase 8690 subunit 727.8 H (EC 3.6.3.14) 274.9352(V-ATPa 61o12 Hypothetical 2330 protein ygaF 8690 620.5389 302.3055 61a24 Protein C20orf24 2560 homolog 8690 615.8312 253.7037 61c24 unknown 2800 8690 670.1639 343.8796 61e24 Voltage-gated 3020potassium 8680channel 1002.349 beta-2255.6019 subunit (K(+) 61g24 hypothetical 3260 protein LOC550520 8690 790.1502 [Danio rerio] 303.2963 61i24 Unclassifiable 3490 EST 8690 877.9728 337.6204 61k24 PREDICTED: 3770similar to 8670 adenylate 822.9764 kinase316.0185 5 isoform 1 [Dan 61m24 Unclassifiable 3970EST 8680 747.2612 289.8148 61o24 Unclassifiable 4200EST 8680 1418.041 354.5648 84a12 RXRgamma? 4430 8680 595.088 305.9445 84c12 buffer 4670 8680 545.9477 228.4444 4900 8680 524.8138 248.9259 5140 8680 556.7717 273.9722 5370 8680 697.3549 299.3148 5600 8680 633.922 260.9445 5840 8680 579.86 267.9908 6080 8680 672.7692 247.2037 6310 8680 738.2451 267.5092 60g12 unknown 2100 8980 837.2375 337.4908 60i12 unknown 2320 8970 782.3333 333.6759 60k12 60S ribosomal 2550protein8970 L6 (TAX-responsive 906.7733 314.9352 enhancer elem 60m12 Unclassifiable 2790EST 8970 775.3107 329.3704 Slide 39 Basic Microarray Data Analysis • Basic information – Channel 1 background and foreground intensity: B1, F1 – Channel 2 background and foreground intensity: B2, F2 • Because two-channel microarray typically use the red and green dye for the two channels, we have: Rb, Rf, Gb, Gf • Background correction R R f Rb G G f Gb • Within-slide normalization R • Identification of differentially M log 2 log 2 R log 2 G G expressed genes A log 2 RG (log 2 R log 2 G) / 2 – Expectation of M is zero. – A gene is overexpressed (or underexpressed) if its M is significantly greater (or smaller) than 0 Xuhua Xia Slide 40 Routine analysis ODS GRAPHICS / ANTIALIASMAX=5800; Data WuduPGF7; Input X Y Ch1I Ch1B Ch2I Ch2B; R=Ch1I - Ch1B; G=Ch2I - Ch2B; if R>0 and G>0 then do; M=log2(R/G); A=log2(R*G)/2; end; else do; M=.; A=.; end; cards; 2280 8520 469.22876 166.962967 2510 8500 443.9776 116.055557 .. ; proc sgplot; scatter x=A y=M ; Xuhua Xia run; 231.892853 295.28302 54.888889 73.981483 Slide 41 MA plot 6 5 4 3 M 2 1 0 -1 -2 -3 3 5 7 9 11 13 15 A Xuhua Xia Slide 42 Microarray data: Background correction /*partial data from slide PGF7*/ Data WuduPGF7; Input X Y Ch1I Ch1B Ch2I Ch2B; cards; 2280 8520 469.22876 166.962967 231.892853 54.888889 2510 8500 443.9776 116.055557 295.28302 73.981483 2750 8500 421.165527 133.40741 227.327103 74.953705 2990 8510 401.165405 151.361115 223.877365 53.722221 ... ; proc g3d; scatter X*Y=Ch1B; run; Run the WuduPGFA.sas file proc loess data=WuduPGF7; model Ch1B=X Y/smooth=0.01 to 0.04 by 0.01 dfmethod=exact degree=2; run; Xuhua Xia Slide 43 Two different kinds of CLM ods output OutputStatistics=StatOut FitSummary=OutSummary; proc loess data=WuduPGF7; model M=A / r CLM smooth = 0.09 alpha=0.01 dfmethod=exact; run; symbol1 c=black i=none value=dot; symbol2 c=black i=join value=none; symbol3 c=red i=join value=none; symbol4 c=red i=join value=none; proc sort data=StatOut out = StatOut2; by A; run; proc gplot data=StatOut2; format DepVar 4.0; plot (DepVar Pred UpperCl LowerCL)*A/ overlay; run; proc reg data=StatOut; model Residual=A / R CLM alpha = 0.01 CLI ; plot Residual *A / pred ; run; Xuhua Xia Slide 44 CLM of the mean Xuhua Xia Slide 45 CLM of the observations Channel-1 background: PGF Slide 7 Ch1B 7328 4899 2469 26230 20260 Y 39 20250 14290 14240 Xuhua Xia X 8230 2220 8320 Slide 47 Channel-2 background: PGF Slide 7 Ch2B 5898 3936 1974 26230 20260 Y 11 20250 14290 14240 X Xuhua Xia 8230 2220 8320 Slide 48 Channel-1 background: 1720p Slide 1 Ch1B 13096 8766 4435 26430 20437 Y 104 20070 14443 14063 X Xuhua Xia 8057 2050 8450 Slide 49 Channel-2 background: 1720p Slide 1 Ch2B 12003 8013 4023 26430 20437 Y 33 20070 14443 14063 X Xuhua Xia 8057 2050 8450 Slide 50 Summary output Number of Observations Number of Fitting Points kd Tree Bucket Size Degree of Local Polynomials Smoothing Parameter Points in Local Neighborhood Residual Sum of Squares Trace[L] GCV AICC AICC1 Delta1 Delta2 Equivalent Number of Parameters Lookup Degrees of Freedom Residual Standard Error 5776 5776 5776 1245 480 480 11 23 34 2 2 2 0.01 0.02 0.03 57 115 173 179605778 194043883 196730653 426.43824 194.50014 166.10841 6.27601 6.22871 6.2512 11.50467 11.49221 11.49548 66457 66381 66400 5314.57324 5560.04084 5570.37194 5372.63763 5571.93922 5544.77939 391.44973 173.04112 126.58875 5257.13638 5548.16787 5596.0826 183.83405 186.81467 187.929 Which smoothing parameter is the best? Xuhua Xia Slide 51 Fit background surface data PredGrid; do Y = 8320 to 26230 by 230; do X = 2220 to 20250 by 230; output; end; end; ods output ScoreResults=Ch1Bscore; proc loess data=WuduPGF7; model Ch1B=X Y/smooth=0.02 dfmethod=exact; score data=PredGrid; run; proc g3d data=Ch1Bscore; format X f4.0; format Y f4.0; format p_Ch1B f4.1; plot X*Y=p_Ch1B/ tilt=60 rotate=80; run; Xuhua Xia Slide 52 Fitted channel-1 background: PGF Slide7 Predicted Ch1B 941 658 26E3 2E4 375 Y 14E3 91.4 2E4 14E3 X 8200 2220 8320 SAS statements for Channel-2 /* With iterative reweighting */ proc loess data=WuduPGF7; model Ch1B=X Y/smooth=0.01 dfmethod=exact ITERATIONS=5; score data=PredGrid; run; proc g3d data=Ch1Bscore; format X f4.0; format Y f4.0; format p_Ch1B f4.1; plot X*Y=p_Ch1B/ tilt=60 rotate=80; run; ods output OutputStatistics=Wudustats; proc loess data=WuduPGF7; model Ch1B=X Y/smooth=0.01 dfmethod=exact ITERATIONS=5; run; proc g3d; scatter X*Y=Pred; run; Xuhua Xia Slide 54 Fitted channel-1 background: PGF Slide7 with ITERATIONS = 5 Predicted Ch1B 250 197 26E3 2E4 144 Y 14E3 90.8 2E4 14E3 X 8200 2220 8320 Fitted channel-1 background: PGF Slide7 with ITERATIONS = 5 Predicted Ch1B 251.77706 197.44784 143.11863 26230 20260 Y 88.78942 20250 14290 14240 X 8230 2220 8320