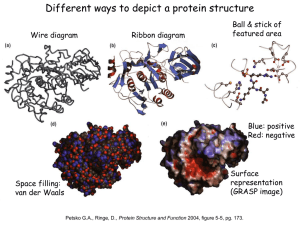
Question 1: Name, PDB codes
advertisement
Question 1: Name, PDB codes • Eric Martz • emartz@microbio.umass.edu • Keiichi Namba • Macromolecular visualization I chose: • 3onz • HUMAN TETRAMERIC HEMOGLOBIN: PROXIMAL NITRITE LIGAND AT BETA • Function: Oxygen transport • Resolution: 2.09 Å • R = 0.218 • Rfree = 0.280 “WORSE THAN AVERAGE at this resolution” Minamino-san gave to me: • 1d66 • DNA-binding domain of Gal4. • Transcriptional regulation. • Resolution: 2.70 Å • R = 0.230 • Rfree not published Question 2: Number of chains Protein: 2 chains (2 distinct) DNA: 0 chains RNA: 0 chains Chain A: HEMOGLOBIN SUBUNIT ALPHA 141 residues (1 missing) of Protein. Source: Homo sapiens (Human). Other_details: Blood. Chain B: HEMOGLOBIN SUBUNIT BETA 146 residues (12 missing) of Protein. Source: Homo sapiens (Human). Other_details: Blood. Question 3: Ligands and non-standard residues 3orn: 2 HEM: Protoporphyrin ix containing Fe 2 NO2: Nitrite ion 4 MBN: Toluene 1d66: 4 CD : Cadmium ion Question 4: Full length vs. crystallized sequence Chain A: Crystal: 146 amino acids Full length: 147 amino acids Chain B: Crystal: 141 amino acids Full-length: 142 amino acids Question 5: Secondary structure This structure is all alpha helices (red) and “coil” (white). It has no beta strands. 80.7% alpha helices 0% beta strands 19.3% neither Question 6: Charge distribution I found no patches of all positive or all negative charges. For 3onz chain A: pI = 8.69. Charges at pH • 4.0: +20.7 • 7.0: +4.3 • 10.0: -7.0 Question 7: Local uncertainty Glu43 in chain B has a high temperature. Question 8 - Hydrophobic cores Yes. Each of the two chains in 3onz has a hydrophobic core (circled in red). Question 9 - Water solubility 3onz appears to be soluble because there are polar residues everywhere on the surface. 3onz is hemoglobin which is known to be a soluble protein. Question 10 - Disulfide bonds Question 11 - Missing residues 13 Missing Residues including 2-, 3+ charged amino acids! All sidechains are complete. Question 12 - Non-covalent interactions • Top: hydrophobic van der Waals interaction between two carbon atoms. • Bottom: histidine nitrogen interacting with negatively charged oxygen (carboxyl). Partial salt bridge since His will have partial positive charge at pH 7, or hydrogen bond since His N epsilon has a hydrogen to donate.. Question 13 - Biological unit Asymmetric unit: 2 chains Biological unit: 4 chains Question 14 - Evolutionary Conservation http://consurf.tau.ac.il/results/1368001022/output.php Top: Lys 61 unexpected. Bottom: Lys 66 expected because it forms salt bridges with the carboxyls on the heme ligand. Question 15 - Proteopedia Scene I Sandbox Reserved 20 (1d66) Question 16 - Proteopedia Scene II Sandbox Reserved 20 (1d66) Question 17a - Cation-pi interaction There are no significant cation-pi interactions in 3onz. 1d66 contains 3 significant cation-pi interactions. One is Lys25 with Tyr40 in chain B. See next slide for snapshot. Here is the report from CaPTURE: Question 17b - Cation-pi interaction (see previous slide for explanation) Question 18 - Polyview-3D static image This image is for 1d66. Deoxyguanosine 26 in chain E is red. Question 19 - Polyview-3D animation for Powerpoint This animation is for 1d66. Deoxyguanosine 26 in chain E is red. Question 20a - Intrinsically disordered regions 3onz chain B is 141 amino acids. The full length sequence has 142. Below are results for the full length sequence. FoldIndex predicts no intrinsically disordered regions, despite the presence of 3 segments of missing residues in the crystal model. Therefore I analyzed 2gry which is more interesting (next slide). Question 20b - Intrinsically disordered regions 2GRY is an X-ray crystallographic structure with resolution 2.35 Å. The full length sequence is 679 amino acids. The crystal includes residues 126-526 (plus an N-terminal His tag). Thus, 1-125 and 527-679 (length 153) were removed before crystallization. FoldIndex predicts that the removed portions are intrinsically disordered. Continued on next slide … Question 20c - Intrinsically disordered regions FoldIndex predicts that 5 segments of the crystallized sequence will be disordered. Segments predicted to be disordered: 1. All but 7 of these 69 N terminal residues are missing in 2GRY. 2. The middle 4 of these 6 residues are missing in 2GRY. 3, 4, 5: None of these predictions overlap with the four additional segments of missing residues in 2GRY. Continued on next slide … Question 20d - Intrinsically disordered regions The two segments of amino acids missing in 2GRY are at the N terminus and near the N terminus. The flanking residues (marked with yellow halos) have mostly low temperatures, while other missing segments (not predicted by FoldIndex) have higher temperatures. 2GRY colored by temperature showing “empty baskets”, regions with missing residues. Yellow halos mark the alpha carbons flanking missing residues predicted to be disordered by FoldIndex.