A 1
advertisement
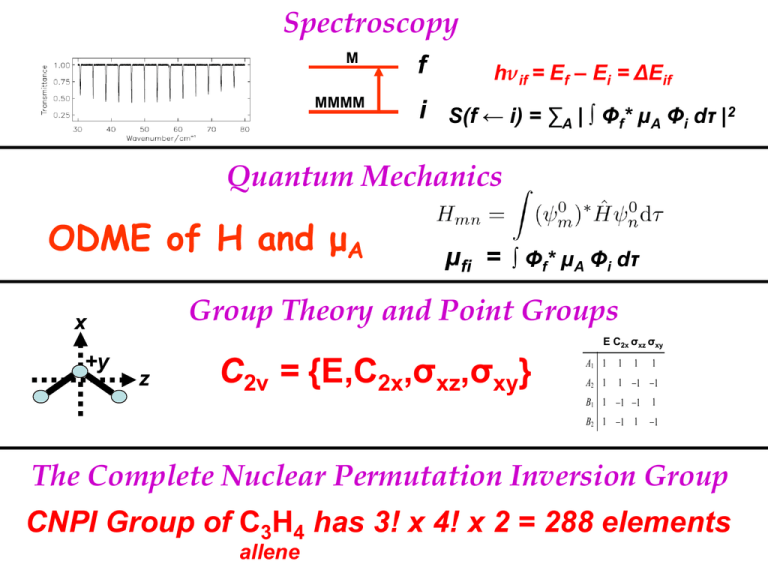
Spectroscopy
M
MMMM
f
hνif = Ef – Ei = ΔEif
i S(f ← i) = ∑A | ∫ Φf* μA Φi dτ |2
Quantum Mechanics
ODME of H and μA
μfi = ∫ Φf* μA Φi dτ
Group Theory and Point Groups
x
σxy
(12)
EE C
E*xz(12)*
2x σ
+y
z
C2v = {E,C2x,σxz,σxy}
A1 1
1
1
1
A2 1
1 1 1
B1 1 1 1
1
B2 1 1 1
1
The Complete Nuclear Permutation Inversion Group
CNPI Group of C3H4 has 3! x 4! x 2 = 288 elements
allene
Labelling Energy Levels Using CNPIG Irreps
R2 = E
RH = HR
A1
A2
B1
B2
E
1
1
1
1
R = ±
(12) E*
1
1
1
-1
-1
-1
-1
1
(12)*
1
-1
1
-1
For example:
Eψ=+1ψ
(12)ψ =+1ψ
E*ψ=-1ψ
(12)*ψ=-1ψ
ψ is a wavefunction of A2 symmetry
∫ΨaHΨbψdτgenerates
= 0 if symmetries of Ψa and Ψb are different.
the A2 representation
∫ΨEIGENFUNCTIONS
of product isIRREDUCIBLY
not A1
aμΨbdτ = 0 if symmetry
TRANSFORM
A2 x B1 = B2, B1 x B2 = A2, B1 x A2 x B2 = A1
The Vanishing Integral Theorem
∫f(τ)dτ = 0 if symmetry of f(τ) is not A1
A1
A2
B1
B2
E
1
1
1
1
(12) E*
1
1
1
-1
-1
-1
-1
1
(12)* Symmetry of H
1
-1
Symmetry of μA
1
-1
Using symmetry labels and the vanishing
integral theorem we deduce that:
∫Ψa*HΨbdτ = 0 if symmetry of Ψa*HΨb is not A1,
i.e., if the symmetry of Ψa is not the same as Ψb
∫Ψa*μAΨbdτ = 0 if symmetry of ψa*μAψb is not A1,
i.e., if the symmetry of the product ΨaΨb is not = symm of μA
A1
A1
A2
B1
Ψ1
0
Ψ2
0
Ψ3
0
Ψ4
0
Ψ5
0
Ψ6
Ψ70
ψ80
B1
ψ1ψ2ψ3ψ4ψ5ψ6ψ7ψ8
. . .
. . .
. . .
. . .
. . .
. . .
. .
. .
0
0
A2
0
0
0
0
0
0
0 0
0
0
0 0
0
A1
A1
A2
B1
Ψ1
0
Ψ2
0
Ψ3
0
Ψ4
0
Ψ5
0
Ψ6
Ψ70
ψ80
B1
ψ1ψ2ψ3ψ4ψ5ψ6ψ7ψ8
. . .
. . .
. . .
. . .
. . .
. . .
. .
B
. 1.
0
0
A2
0
0
0
0
0
0
A1 0 0
0 A2 0
0 0
0
Allowed
Transitions
A 1 ↔ A2
B1 ↔ B2
Connected
by A2
Using the CNPI Group
for any molecule
Like using a big computer to calculate
E from exact H, we can, in principle, use
the CNPI Group to label energy levels
and to determine which ODME vanish
for any molecule
BUT there is one big problem:
Superfluous symmetry labels
Using the CNPI Group
approach for CH3F
Character Table of CNPI group of CH3F
GGCNPI
CNPI
12 elements
6 classes
6 irred. reps
E (123) (23) E* (123)* (23)*
(132) (31)
(132)* (31)*
(23)
(23)*
H A1’
μA A1”
CH3F
The CNPI Group approach
E’ + E’’
A1’
E’ + E’’
A1’’
A1’’ + A2’
A 2’
A2’’
A1’ + A2’’
E’
E’’
Block diagonal H-matrix
Allowed
transitions
connected
by A1”
CH3F
The CNPI Group approach
E’ + E’’
A1’
E’ + E’’
A1’’
A1’’ + A2’
A 2’
A2’’
A1’ + A2’’
E’
Allows for tunneling
E’’between two
VERSIONS
CH3F
TWO VERSIONS
F
Very, very
high
potential
barrier
F
2
3
1
2
1
3
No observed tunneling through barrier
The number of versions of the minimum is given by:
(order of CNPI group)/(order of point group)
For CH3F this is 12/6 = 2
3! x 2
C3v group has 6 elements
For benzene C6H6 this is 1036800/24 = 43200,
and using the CNPI group each energy level would
get as symmetry label the sum of 43200 irreps.
Clearly using the CNPI group gives
very unwieldy symmetry labels.
CH3F
Only NPI OPERATIONS FROM IN HERE
F
Very, very
high
potential
barrier
F
2
3
1
2
1
3
(12) superfluous
E*
superfluous
(123),(12)* useful
No observed tunneling through barrier
If we cannot see any effects
of the tunneling through the
barrier then we only need
NPI operations for one
version. Omit NPI elements
that connect versions since
they are not useful; they are
superfluous
superfluous.
For CH3F:
superfluous
useful
GCNPI={E, (12), (13), (23), (123), (132),
E*, (12)*, (13)*,(23)*, (123)*, (132)*}
useful elements are
The six feasible
GMS ={E, (123), (132), (12)*, (13)*,(23)*}
Character table of the MS group of CH3F
(12),(13),(23)
E*,(123)*,(132)*
are unfeasible A1
elements of the
A2
CNPI Group
E
E (123) (12)*
E (123) (12)
(132) (13)*
1
2 (23)*
3
1
1
1
1
1
1
2
1
0
H A1
μA A2
Use this group to block-diagonalize H
and to determine which transitions are
forbidden
Using CNPIG versus MSG for PH3
E’ + E’’
E
E’ + E’’
E
A1’’ + A2’
A2
A1’ + A2’’
A1
CNPIG
MSG
Can use either to determine if an ODME vanishes.
But clearly it is easier to use the MSG.
Superfluous
Unfeasible elements of the CNPI group
interconvert versions that are separated
by an insuperable energy barrier
useful
The subgroup of feasible elements forms a group called
THE MOLECULAR SYMMETRY GROUP
(MS GROUP)
----------- End of Lecture Two -------
16
We are first going to set up the
Molecular Symmetry Group for
several non-tunneling (or “rigid”)
molecules. We will notice a strange
“resemblance” to the Point Groups
of these molecules. We will examine
this “resemblance” and show how it
helps us to understand Point Groups.
x
H2O
(+y)
1
z
2
C2v(M) elements:
C2v elements:
{E, (12), E*, (12)*}
{E
C2x σxz
σxy}
The C2v and C2v(M) character tables
x
H2O
(+y)
1
C2v(M)
z
2
E (12) E* (12)*
C2v
E (12)
C2x E*
σxz (12)*
σxy
E
A1
1
1
1
1
A1
1
1
1
1
A2
1
1
1
1
A2
1
1
1
1
B1
1
1 1
1
B1
1
1 1
1
B2
1
1
1
B2
1
1
1
1
1
The C3v and C3v(M) character tables
F
3
CH3F
2
1
C3v(M)
E (123) (12)*
(123) (13)*
(12)
E (132)
1
2 (23)*
3
C3v
E C3 σ1
(123)
(12)
E C
2 σ
3
2
1
2 σ33
A1
1
1
1
A1
1
1
1
A2
1
1
1
A2
1
1
1
E
2
1
0
E
2
1
0
HN3 has
6 versions
12/2 = 6
3!x2
Cs
H
N1 N2 N3
GCNPI={E, (12), (13), (23), (123), (132),
E*, (12)*, (13)*,(23)*, (123)*, (132)*}
All P and P* are unfeasible (superfluous)
MSG is {E,E*}
E
A’ 1
A” 1
E*
1
-1
HN3 has
6 versions
12/2 = 6
3!x2
Cs
H
N1 N2 N3
GCNPI={E, (12), (13), (23), (123), (132),
E*, (12)*, (13)*,(23)*, (123)*, (132)*}
All P and P* are unfeasible (superfluous)
MSG is {E,E*}
E
A’ 1
A” 1
E*
1
-1
PG is {E,σ}
E
A’ 1
A” 1
σ
1
-1
H3
+
1
+
2
3
GCNPI={E, (12), (13), (23), (123), (132),
E*, (12)*, (13)*,(23)*, (123)*, (132)*}
Point group also has 12 elements: D3h
Therefore only 1 version and MSG = CNPIG
Character Table of MS group H3+
GCNPI
GCNPI
12 elements
6 classes
6 irred. reps
E (123) (23) E* (123)* (23)*
(132) (31)
(132)* (31)*
(23)
(23)*
Character table for D3h point group
linear,
E 2C3 3C'2 σh 2S3 3σv
q
rotations
A'1 1 1 1
1 1 1
x
A'2 1 1 -1 1 1 -1 Rz
E' 2 -1 0
2 -1 0 (x, y)
(
A''1 1 1 1
-1 -1 -1
A''2 1 1 -1 -1 -1 1 z
E'' 2 -1 0
-2 1 0 (Rx, Ry) (
The allene molecule C3H4
Number of elements in CNPIG = 3! x 4! x 2 = 288
H5
H4
H7
C1
C2
Point group is D2d has 8 elements
Thus there are 288/8 = 36 versions
C3
H6
The MSG of allene is:
{E, (45)(67), (13)(46)(57), (13)(47)(56), (45)*, (67)*,
(4657)(13)*, (4756)(13)*}
The allene molecule C3H4
Number of elements in CNPIG = 3! x 4! x 2 = 288
H5
H4
H7
C1
C2
Point group is D2d has 8 elements
Thus there are 288/8 = 36 versions
C3
H6
The MSG of allene and PG are:
E
C2
C2’
C2 ’
σd σd
{E, (45)(67), (13)(46)(57), (13)(47)(56), (45)*, (67)*,
S4
S4
(4657)(13)*, (4756)(13)*}
Character table for D2d point group
linear,
E 2S4 C2 (z) 2C'2 2σd
rotations
A1 1 1 1
1
1
A2 1 1 1
-1 -1 Rz
B1 1 -1 1
1
-1
B2 1 -1 1
-1 1 z
E 2 0 -2
0
0 (x, y) (Rx, Ry)
29
USE OF GROUP THEORY AND
SYMMETRY IN SPECTROSCOPY
Use
molecular
geometry
Point Group
Spoilt by rotation
and tunneling
Use energy
invariance
MS Group
Superfluous
symmetry if
many versions
RH = HR
CNPI Group
RH=HR, therefore
can symmetry label
energy levels
USE OF GROUP THEORY AND
SYMMETRY IN SPECTROSCOPY
Use
molecular
geometry
Point Group
Spoilt by rotation
and tunneling
Use energy
invariance
MS Group
Superfluous
symmetry if
many versions
RH = HR
CNPI Group
RH=HR, therefore
can symmetry label
energy levels
USE OF GROUP THEORY AND
SYMMETRY IN SPECTROSCOPY
Use
molecular
geometry
Point Group
MS Group
Spoilt by rotation
and tunneling
Use energy
For rigid
(nontunneling)
invariance RH = HR
molecules MSG and PG
are isomorphic. Leads to
CNPI
Group
an
understanding
of
PGs
Superfluous
RH=HR,
therefore
symmetry
if they
and how
can
label
can symmetry label
many versions
energy levels.
energy levels
Black are instantaneous
positions in space.
White are equilibrium
positions.
N.B. +z is 1→2.
Then do (12).
Note that axes have moved.
Rotational coordinates are
transformed by MS group.
Black are instantaneous
positions in space.
White are equilibrium
positions.
N.B. +z is 1→2.
Then do (12).
Note that axes have moved.
Rotational coordinates are
transformed by MS group.
Undo the permutation
of the nuclear spins
Undo the permutation
of the nuclear spins
We next undo the
Rotation of the axes
only ev
coords
only ev
coords
only rot
coords
only nspin
coords
MS Group and Point Group of
H2 O
C2v(M)
MS Group
E =
Point Group
p0 R0 E
(12) = p12 Rx C2x
E* = p0 Ry xz
(12)* = p12 Rz xy
C2v
MS Group and Point Group of
H2 O
C2v(M)
MS Group
E =
Point Group
p0 R0 E
(12) = p12 Rx C2x
E* = p0 Ry xz
(12)* = p12 Rz xy
C2v
Jon Hougen
and what the
MSG is called
by some
people
Point group operations rotate/reflect
electronic coords and vib displacements
in a non-tunneling (rigid) molecule.
RPG Hev = Hev RPG
The point group can be used to symmetry
label the vibrational and electronic
states of non-tunneling molecules.
The rotational and nuclear spin
coordinates are not transformed
by the elements of a point group
47
The MSG can be used to classify
The nspin, rotational, vibrational
and electronic states of any
molecule, including those
molecules that exhibit tunneling
splittings (“nonrigid” molecules).
WE USE THE ETHYL RADICAL AS AN
EXAMPLE OF A NONRIGID MOLECULE
The ethyl
radical
H4
a
Cb
H5
Number of elements in CNPIG = 5! x 2! x 2 = 480
The MSG of rigid (nontunneling) ethyl is {E,(23)*}
The MSG of internally rotating ethyl is the
following group of order 12:
{E, (123),(132),(12)*,(23)*,(31)*,
(45),(123)(45),(132)(45),(12)(45)*,(23)(45)*,(31)(45)*}
There are still plenty of unfeasible elements
such as (14), (23), (145), E*, etc.
MS group can change with
resolution and temperature
THE PH3 MOLECULE AS AN EXAMPLE
Barrier height = 12300 cm-1
Schwerdtfeger, Laakkonen and Pekka Pyykkö,
J. Chem. Phys., 96, 6807 (1992)
Recent theoretical calcs by
Sousa-Silva, Polyanski, Yurchenko and Tennyson
From UCL, UK.
Yield the tunneling splittings given on the next slide
7.2 cm-1
0.775
0.145
0.023
0.0017 cm-1
15 MHz
2 MHz
240 kHz
15 kHz
600 Hz
12 Hz
D3h(M)
CNPIG
C3v(M)
Tunneling splittings in NH3
D3h(M)
MS group can change
with resolution
and temperature
For PH3 MSG is usually C3v(M)
For NH3 MSG is usually D3h(M)
SUMMARY
54
Unfeasible elements of the CNPI group
interconvert versions that are separated
by an insuperable energy barrier
The subgroup of feasible elements form a group called
THE MOLECULAR SYMMETRY GROUP
MS group can change with resolution and T
MS group is used to symmetry label the
rotational, ro-vibrational, rovibronic and
nuclear spin states of any molecule.