Slide 1
advertisement

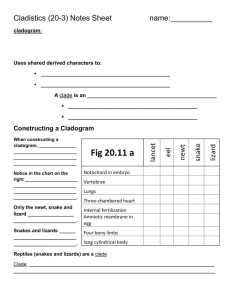
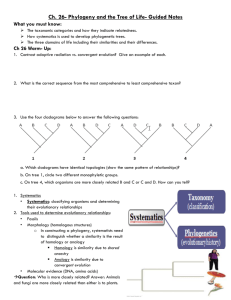
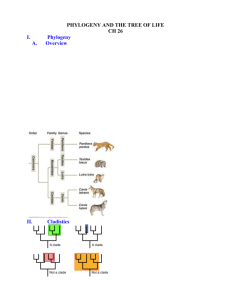
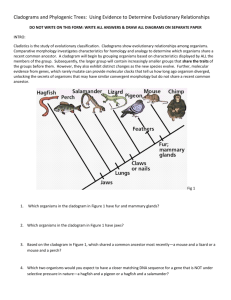
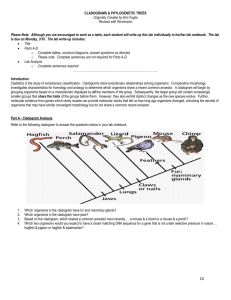
Phylogenetic Trees - I Phylogenetic Tree - phylogenetic relationships are normally displayed in a tree-like diagram (phylogenetic tree/cladogram) - a cladogram is a branching diagram describing the phylogenetic relationships of the taxa under investigation. - a phylogenetic tree graphically represents a hypothetical evolutionary process. - phylogenetic trees are subjected to revision as additional data becomes available. - evolutionary tree is used synonymously as phylogenetic tree. Phylogenetic Tree - a cladogram consists of two major elements: nodes and branches. - a node represents a taxon; a branch represents evolutionary event(s). - the branching pattern of a cladogram is called the topology. Phylogenetic Trees Cladogram Clade - a clade is an ancestor species with all its descendants. - a monophyletic group is a clade. - There are three ways to define a clade for use in a cladistic taxonomy i) Node based ii) Stem based iii) Apomorphy based Clade Node-based the most recent common ancestor of A and B along with all of its descendants Stem-based all descendants of the oldest common ancestor of A and B that is not also an ancestor of Z Apomorphy-based the most recent common ancestor of A and B, along with all of its descendants, possessing a certain derived character. This definition is generally discouraged by most cladists Cladogram / Phylogram / Phenogram - A cladogram/phylogram displays branching information to explain the evolutionary relationships. - A phylogram has additional information: the length of branches according to the amount of changes (evolutionary process). Cladogram / Phylogram Cladogram Phylogram Cladogram / Phylogram / Phenogram - A phenogram is generated from phenetic analysis (numerical taxonomy). It does not necessary explain the ancestor-descendant relationships of the taxa in the investigation. Gene trees and species trees Gene tree a A b B c C Species tree We often assume that gene trees give us species trees Characters and Character States • Organisms comprise sets of features • When organisms/taxa differ with respect to a feature (e.g. its presence or absence or different nucleotide bases at specific sites in a sequence) the different conditions are called character states • The collection of character states with respect to a feature constitute a character Homoplasy • Homoplasy is similarity that is not homologous (not due to common ancestry) • It is the result of independent evolution (convergence, parallelism, reversal) • Homoplasy can provide misleading evidence of phylogenetic relationships (if mistakenly interpreted as homology) • homoplasy can be common in DNA data – There are a limited number of alternative character states (e.g. Only A, G, C and T in DNA) – Rates of evolution are sometimes high