RFA-HG-14-001 Applicant Information Webinar
advertisement
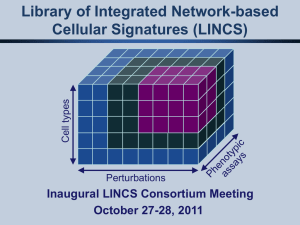
BD2K-LINCS-Perturbation Data Coordination & Integration Center Applicant Information Webinar for RFA-HG-14-001 Ajay Pillai and Jennie Larkin January 13, 2013 1:00 - 2:30 PM EDT RFA-HG-14-001 Applicant Information Webinar BD2K-LINCS-Perturbation Data Coordination and Integration Center (DCIC) (U54) Today’s Webinar: • BD2K and LINCS program introduction • Overview of new FOA • Questions Big Data To Knowledge (BD2K): Overview A trans-NIH initiative BD2K Mission enable biomedical scientists to capitalize more fully the Big Data being generated by the research community http://bd2k.nih.gov/ BD2K: Background • Major challenges in using biomedical Big Data include: – – – – Locating data and software tools. Getting access to the data and software tools. Standardizing data and metadata. Extending policies and practices for data and software sharing. – Organizing, managing, and processing biomedical Big Data. – Developing new methods for analyzing & integrating biomedical data. – Training researchers who can use biomedical Big Data effectively. BD2K Centers • There was a separate call for Investigator-initiated Centers (RFA-HG-13-009) • This will be the first NIH-specified BD2K center. • This center will focus on perturbation – response data, including that generated by the LINCS consortium. • This Center will include the BD2K focus areas: – Collaborative environments and technologies – Data Integration LINCS aims to inform a network-based understanding of biological systems in health and disease that can facilitate drug and biomarker development. LINCS is: Developing a library of molecular and cellular signatures that describe how different cell types respond to a variety of perturbations. Addressing challenges in high-throughput data generation, data integration, annotation, and analysis. Actively exploring collaborations with new biomedical research communities. Human cell types LINCS: Library of Integrated Network-based Cellular Signatures Perturbations • RNAi • small molecules http://lincsproject.org LINCS Program (2014 – 2020) • LINCS goals – inform a network-based understanding of cellular functions and response – expand the scope and richness of cellular responses to be measured. – support the addition of a broader and more informative range of human cell types, perturbations, and measurements. • LINCS Program Structure – 3-5 Data and Signature Generating Centers (RFA-RM13-013) to be funded in FY14 – One BD2K-LINCS Perturbagen Data Coordination and Integration Center (RFA-HG14-001) to be funded in FY15 – 6 year program with Mid-Course Review (~July 2017) Background: LINCS Data and Signature Generating Centers • Data and Signature production at scale, within first year of award (tens of thousands of data points per year) • Cell Types: human cells (cell lines, primary tissue, iPS cells and their differentiated derivatives) • Perturbagens: – Pilot: small molecules, growth factors, and genetic (knockdown or upregulation by gene overexpression) – These will continue but applicants may propose other perturbations • Assays: – – – – Should be medium to high throughput Provide measures of wide interest to biomedical researchers Should be flexible and amenable to multiple cell types Should be replicable with high level of QC/QA under SOPs BD2K-LINCS Perturbagen DCIC HG-14-001 • Aims in both section I and IV of RFA: read both carefully • 1 award, $5M in 2015. Future year amounts will depend on annual appropriations. • Application budget may be up to $3 million direct costs per year, not including the F&A costs of subcontracts. • 5-yr duration, it is a cooperative agreement • Familiarize yourself well with RFA-RM-13-013 • Data science is described in RFA-HG-13-009. BD2K-LINCS Perturbagen DCIC Goals • address significant data science challenges associated with perturbagen-response datasets • establish a community resource for perturbagen-response data • coordinate LINCS consortium activities • Goal: enable advances in understanding of cellular function and its relationship with disease and normal biology BD2K-LINCS Perturbagen DCIC • Integrated Knowledge Environment – Data Integration: • integrating LINCS data with other perturbation data and other non-perturbation datasets – Collaborative Environments and Technologies: • utilize novel methods to provide access while supporting data attribution and provenance – Support Unified Access to LINCS DSGC Resources: • Support single-point of access for community to DSGC and DCIC tools & data • For bench & computational scientists LINCS Data/Signature Access • Each DSGC will build an appropriate database and an underlying infrastructure to support queries and other analytical requirements on their datasets • Metadata annotation by DSGCs for both data and software resources is crucial. • LINCS will have a distributed data resource and infrastructure to support queries • LINCS aims to create a single user interface via the separate DCIC for all of the LINCS resources for all biomedical researchers, including computational biologists BD2K-LINCS Perturbagen DCIC • Data Science Research Collaborations – Internal innovative DSR projects related to perturbation data; short-term; adaptable/flexible; – External Data Science Collaborations: • bring in novel expertise and analytical capabilities, to engage in high-risk high-reward approaches • set aside $700,000 in direct costs each year • identify 3 collaborative projects (lasting 12 months) with groups that are not part of the application • Propose a plan to identify three such innovative projects each year of the funded grant BD2K-LINCS Perturbagen DCIC • Consortium Coordination and Administration – May request up to $100,000/yr for BD2K coordination efforts – Support Incorporation of LINCS-related Data Types from External Resources • You do not expected to replicate other databases, but can retain relevant indexes/summaries for efficiency in retrieval – Coordinate Annotation of Data, Tools, and Resources • Enable coordination activities for the LINCS consortium (DSGCs and the DCIC) BD2K-LINCS Perturbagen DCIC • Community Training and Outreach – Data science • address questions of access and use of perturbationtype by community – Access to LINCS Resources • Work with LINCS DSGC to establishing the LINCS resource & approach within multiple biomedical communities. • Propose how your training/outreach will enable subsets of the biomedical community to leverage the whole LINCS resource. DCIC: program administration • Cooperative agreement, with substantial collaboration between LINCS grantees and involvement of program staff. – Integral part of LINCS Steering Committee with relevant and appropriate leadership role to enable overall LINCS goals. – Participate in BD2K Working Groups and other suitable activities including annual BD2K meetings. • Questions: lincsproject@mail.nih.gov DCIC: Review • Reviewers will provide an impact score for each component of the Center; Impact score of the Overall Component is the impact score of the entire application. • Some significant questions: – data integration challenges within and across LINCS & other existing public resources – single user-interface for all LINCS data & signature – community access & scalability – coordination & metadata for LINCS – integration of components of the center APPENDIX NIH Common Fund • Supports cross-cutting programs that are expected to have exceptionally high impact. • Develops bold, innovative, and often risky approaches to address problems that may seem intractable or to seize new opportunities that offer the potential for rapid progress. • NIH LINCS Program Co-Chairs: – Alan Michelson, PhD (NHLBI) – Mark Guyer, PhD (NHGRI) • NIH LINCS Coordinators – Ajay Pillai, PhD (NHGRI) – Jennie Larkin, PhD (NHLBI) LINCS Pilot Phase (2010 – 2013) • Pilot goals: – Develop a limited yet coherent data, and signature resource that could be used by the general research community. – Identify key issues in data annotation, integration, and analysis. • Pilot activities: – Two data and signature generating U54 awards – Development of new high-throughput assays to detect perturbation-induced cellular responses – Novel computational methods for integrative data analysis – Active collaborations and working groups http://lincsproject.org Background: LINCS Data and Signature Generating Centers • • • • RFA-RM13-013 (going to May 2014 Council) Will fund 3-5 DSGC awards Part of a collaborative LINCS program DSGC structure: 1. 2. 3. 4. Data Generation (40% effort) Data Analysis and Signature Identification (40% effort) Community Interactions Outreach (20% effort) Administrative BD2K Centers • A combination of Investigator-Initiated and NIHspecified Centers • Centers to conduct research & provide resources • Centers will form an interactive consortium • Investigator Initiated Centers FOA : Centers of Excellence for Big Data Computing in the Biomedical Sciences (U54) RFA-HG-13-009 – 6-8 will be funded Summer 2014. • Potential Centers focus areas: – – – – Collaborative environments and technologies Data Integration Analysis and modeling methods Computer science and statistical approaches NIH Big Data to Knowledge (BD2K) Programmatic Areas I. Facilitating Broad Use of Biomedical Big Data: Mike Huerta NLM & Jennie Larkin NHLBI II. Developing and Disseminating Analysis Methods and Software for Biomedical Big Data: Vivien Bonazzi NHGRI & Jennifer Couch NCI III. Enhancing Training for Biomedical Big Data: Michelle Dunn NCI IV. Establishing Centers of Excellence for Biomedical Big Data: Lisa Brooks NHGRI, Mike Huerta NLM, Peter Lyster NIGMS & Belinda Seto NIBIB) Perturbation DCIC: linking two programs (BD2K and LINCS) • BD2K: supports necessary advances in data science, other quantitative sciences, policy, and training to support the effective use of Big Data in biomedical research. • LINCS: promote a new understanding of health and disease through an integrative approach that identifies common patterns (signatures) in molecular and cellular responses to a wide range of perturbations, including small molecules, other environmental stimuli, genetic variation, and disease