Enzyme Regulation/Rates PPT
advertisement
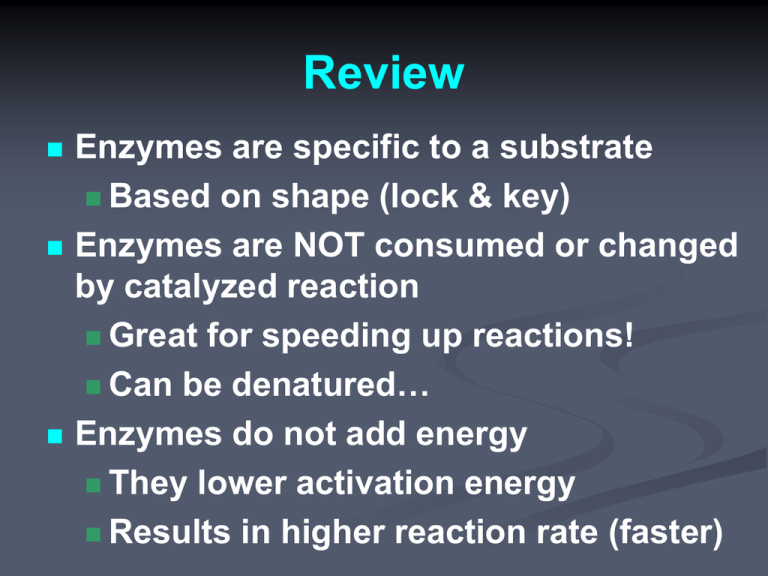
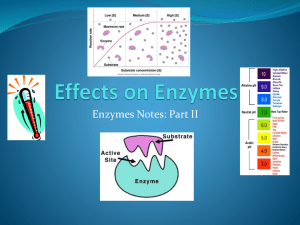
Review Enzymes are specific to a substrate Based on shape (lock & key) Enzymes are NOT consumed or changed by catalyzed reaction Great for speeding up reactions! Can be denatured… Enzymes do not add energy They lower activation energy Results in higher reaction rate (faster) Enzyme Regulation How cell affects enzyme activity (rate) “on purpose” regulate activity Faster/slower? Enzyme on/off ? Can send “signal molecule” binds enzyme in 2 ways: 1) Can bind directly to active site: prevents substrate bind only repress 2) Can bind “elsewhere” on enzyme to change enzyme shape (allow OR prevent Allosteric substrate binding): activate or repress site Allosteric enzymes: can change to other shape by signal molecule Enzyme Regulation Names If repressor signal molecule binds to active site… Called competitive inhibition If repressor signal molecule binds to allosteric site… Enzyme changes shape Active site no longer fits Called noncompetitive inhibition Not all enzymes have an allosteric site Not all enzymes are allosteric… http://www.northland.cc.mn.us/biology/biology1111/animations/enzyme.swf Enzyme Inhibitors http://bcs.whfreeman.com/thelifewire/content/chp06/0602002.html http://course1.winona.edu/sberg/animtns/allostan.gif Allosteric Enzyme Regulation Active Site Repressor signal molecule Will change shape so CANNOT bind Activator signal molecule Will change shape so CAN bind Allosteric Site Feedback Inhibition System of enzyme regulation where product of reaction can be signal molecule (inhibition repressor) Competitive feedback inhibition Product Product Product Product Product binds directly to active site Product Product Product Product Product Noncompetitive feedback inhibition Product Product binds to allosteric site Product Product Product Allows for self-regulation If too much product, stops catalysis (repress) If too little product, continues catalysis Binding not permanent (can diffuse away) Product http://www.northland.cc.mn.us/biology/biology1111/animations/enzyme.swf - Feedback Inhibition (Biochemical) Metabolic Pathway A “chain” reaction Involves several enzymes (closeby) Product of one catalyzed reaction becomes substrate for next Desired product only made at end Similar to Assembly Line of pathway Allows for more precise control http://highered.mcgrawhill.com/classware/ala.do?isbn=0072986670&alaid=ala_1048052&showSelfStudyTree=true http://highered.mcgraw-hill.com/classware/ala.do?isbn=0072986670&alaid=ala_1048055&showSelfStudyTree=true Prepare for a mix/match of different regulations Noncompetitive Inhibition = allosteric enzyme with repressor (simple) Noncompetitive Feedback Inhibition = allosteric enzyme with product being repressor Noncompetitive Feedback Inhibition of a Biochemical Pathway = several enzymes with at least 1st enzyme being allosteric and product of last enzyme being a repressor to 1st enzyme Environmental Affects on Enzymes Increase amount of substrate Increase amount of enzyme Increase temperature Increase pH Increase an repressor molecule Having a Feedback Inhibition Protein Protein works best works best Rate Protein denatures Protein works slow Protein denatures Substrate Enzyme Temperature pHRepressor Time Substrate An Example H2O2 = Hydrogen peroxide (reactant) Can spontaneously break down to H2O water and O2 gas (products)… so this is an… Exergonic reaction 2H2O2 2H2O + O2 Enzymes speed up this reaction by… lowering activation energy…how exactly? In terms of motion of molecules, how is it lowered? Energy Profile of Exergonic Rxn Note release of energy Note EA Activation Energy Energy required to start reaction = The energy needed to get molecules into transition state Can accomplish by moving faster (i.e. add heat: thermal energy) Enzymes lower Activation Energy Release of energy same as without enzyme How enzymes lower activ. nrg? (molecular level) Substrate-Specificity (by shape) Substrate = reactant(s) catalyzed by enzyme Substrate binds to enzyme on active site Forms enzyme-substrate complex (H-bonds) Induced fit: enzyme changes shape SLIGHTLY to “cuddle” substrate Active Site is shapespecific – catalyze on specific substrate Induced fit lowers activation energy because orients substrate(s) correctly Enzyme Catalysis Enzyme does NOT change in catalysis (retains shape) Enzyme is ready to catalyze another substrate after reaction is complete (after products released) Note on Molecular Motion: It’s still all about “chance” Substrates can hit enzyme in “wrong” way and not be catalyzed… But since enzyme has active site (fitted shape), more likely to react than if substrate on own Rate of Catalysis Reaction How quickly turn reactants into products NOT how much products are made Rate of Catalysis = rate of enzymes How quickly do enzymes catalyze Rate is affected by several environmental factors Rate of Catalysis Rate = 2 1 rxn in 1 sec Rate of Catalysis Rate = 10 rxn in 1 sec Rate of Catalysis Rate = 20 rxn in 2 sec = 10 rxn in 1 sec Rate of Catalysis As you increase substrate concentration, rate will increase until… Enzymes are saturated and then rate will level off (enzyme still working…) As you increase enzyme concentration, rate will increase (indefinitely?) Only when Enzymes >>>>> Substrate, rate will level off eventually Saturation more likely if increase substrate (not enzyme) Rate of Catalysis Rate = 20 rxn in 2 1 sec Rate of Catalysis With more enzymes, there is a higher chance of substrate hitting correctly and reacting -- this increases rate But at some point, rate will level off Compared to increasing substrate, this level off will be (@ a much higher concentration of enzyme and @ a much higher rate) Rate = 5 rxn in 0.9 1 sec sec Graphing Enzyme Rates Increase amount of substrate Increase amount of enzyme Rate Enzyme Substrate 2 more factors affecting enzyme rate Same 2 factors that cause denaturation… Temperature Increase temp. will move molecules faster but…break pH H-bonds in enzyme: not functional Change in H+ concentration will disrupt H-bonds affect cross-linking + Environmental Affects on Enzymes Increase amount of substrate Increase amount of enzyme Increase temperature Increase pH Increase an repressor molecule Having a Feedback Inhibition Protein Protein works best works best Rate Protein denatures Protein works slow Protein denatures Substrate Enzyme Temperature pHRepressor Time Substrate