Description
advertisement

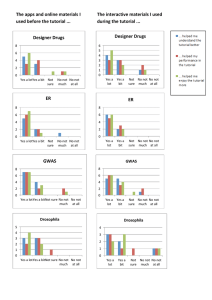
BMI206 Network biology lab Nov 20, 2014 Lab materials (files) • Parent_PPI: This is a highly-curated human protein interaction network • HT.pvals.out: Gene-based p-values (VEGAS) from a GWAS in hypertension (HT) • MS.pvals.out: Gene-based p-values (VEGAS) from a GWAS in multiple sclerosis (MS) • Cytoscape_import_attributes.txt: Gene-based pvalues from both GWAS and known associated genes for importing into Cytoscape Lab materials (code) • qqplot.r: Script to run gene-wise Qqplot from each GWAS • Manhattan plot.r: Script to run gene-wise Manhattan plots from each GWAS • Make_block.r: Script to compute association blocks from both GWAS and plot them side by side • Pathway_permutation.r: script to run background distributions of networks of different sizes with randomly selected nodes from the parent PPI. This code also plots the distributions and the actual number of significant genes (nodes) in the first order nets Lab materials (software) • Cytoscape 3.2 – Network analyzer (Core plugin) – BINGO plugin Assignment 1. After exploring the Manhattan plots, qq-plots and association blocks from each GWAS, what can you tell about the power of each study? 2. Using Cytoscape, analyze the PPI and describe its main network properties 3. Using Cytoscape, find the first order networks (p<0.05) for each GWAS 4. Are the first order networks from both GWAS more connected than expected? What does this mean? 5. Run BINGO App on all nodes. nodes from largest connected component. What biological processes emerge from the first order networks? 6. Run BINGO App on nodes from largest connected component. Interpret the results 7. Map and color known MS and HT genes onto their respective first order nets. Interpret results.