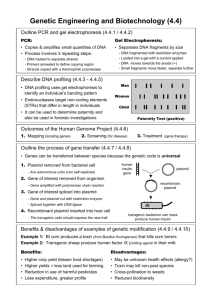
Republic of the Philippines RIZAL TECHNOLOGICAL UNIVERSITY Cities of Mandaluyong and Pasig COLLEGE OF ARTS AND SCIENCES Asynchronous Activity 6 LABORATORY REPORT GROUP 2 MEMBERS: ● BATAC, Bea Adrianne ● BERSUELA, Alexandria M. ● FLORES, Niña Genilou V. ● RAÑA, Christina Anne Mae P. ● RAZON, Angela Mae M. SECTION: CAS – 02 – 501A BS BIOLOGY SUBJECT: CELL AND MOLECULAR BIOLOGY SCHEDULE: MWF 7:30 AM TO 9:00 AM INTRODUCTION Plasmids are small circular DNA molecules, ranging in size from 1,000 to 200,000 base pairs that are present in multiple copies separate from the chromosomal DNA. The basic components of a plasmid are the ori site for initiation of DNA replication, a promoter for the initiation of transcription, and a gene for antibiotic resistance. Plasmid is critical for genetic engineering in which they can be passed on from one bacterial strain to another in a process called bacterial conjugation, which enables bacteria to share and exchange genetic information. Scientists have taken advantage of plasmids to use them as tools to clone, transfer, and manipulate genes. Plasmids that are used experimentally for these purposes are called vectors. Researchers can insert genes of interest into a plasmid vector, creating a so-called recombinant plasmid. This plasmid can be introduced into a bacteria by the process called transformation. Then, because bacteria divide rapidly, they can be used as factories to copy DNA fragments in large quantities.In DNA cloning, restriction enzymes and plasmids are used together to create a recombinant plasmid. A restriction enzyme is a DNA-cutting enzyme that recognizes specific sites in any given DNA sequence. It should cut in two places: (1) on either side of the gene of interest; and (2) within the plasmid backbone. If two DNA molecules have matching ends, it creates compatible sticky ends on both pieces of DNA that can be re-attached to make the recombinant plasmid by the use of enzyme DNA ligase. DNA ligase seals the gap between the molecules, forming a single piece of DNA. When making a recombinant plasmid, it is important to verify that the restriction digest and ligation procedures worked as expected. Through the use of gel electrophoresis, the sizes of the restriction digest fragments and ligation product are being compared from the products of both procedures with each other and with what you started with to ensure that the desired recombinant plasmid was obtained. Hence, to know if you have made the recombinant plasmid that is needed. METHODS The restriction digest was first conducted in building a recombinant plasmid, in this simulation we produce two DNA fragments: one fragment that contained a gene of interest, and the other fragment is a plasmid backbone that is used to express proteins in bacterial cells. These two plasmids are combined by gene cloning in creating recombinant plasmids, the gene cloned is called rfp. In cloning of the rfp gene, DNA from the two plasmids are used, these are the pKAN-R which carries the rfp gene, and the pARA that makes bacteria resistant to antibiotic ampicillin. The second simulation is DNA ligation, this is where the DNA fragments resulting from the restriction digest of pKAN-R and pARA are joined to create the recombinant pARA-R plasmid. For the first and second simulation we separately digest (cut) the gene fragment and the plasmid. This step produces fragments with sticky ends. Next, we take the gene fragment and the linearized (openedup) plasmid and combine them along with DNA ligase. The sticky ends of the two fragments stick together by complementary base pairing. Once they are joined by ligase, the fragments become a single piece of unbroken DNA. The target gene has now been inserted into the plasmid, making a recombinant plasmid. Last is the laboratory simulation of verifying a recombinant plasmid by gel electrophoresis, there are 10 steps that were completed in order to obtain an ideal result. First, the protocol table was filled up with needed information about the volumes of each reagent required to add in the experiment. Second is the addition of H2O to the sample tubes. With the use of P20 micropipette, a 4μl distilled water (dH₂O) was added to gK− tube, this procedure was repeated in the tubes of gK+, gA−, and gA+, while for the gLIG tube a 3µl of dH₂O was added instead of 4µl. Third step is the addition of loading dye to the sample tubes, using a P2 micropipette a 2μl Loading Dye (LD) was added in gK− tube, this procedure was repeated for the tubes of gK+, gA−, gA+ and gLIG. Fourth is the addition of samples to the sample tubes, a P20 micropipette was used to add 4μl of Ksolution to the gK− tube, this procedure was also repeated to gK+, gA−, gA+, and gLIG tube. Fifth step is the preparation of the microcentrifuge, each of the solutions mentioned above was moved to the microcentrifuge and was briefly spinned. Next is the preparation of the electrophoresis box where an agarose gel was placed inside of it, and a sodium borate buffer (1XSB) was poured over the gel. Seventh step is the DNA Ladder draw up, a P20 micropipette was used to add 10μl of DNA Ladder (M) to the lane 1 of the gel in the electrophoresis box. After this the gK− and other solutions was also pipetted into the other wells, gK− solution into lane 2, gK+ solution into lane 3, gA− solution into lane 4, gA+ solution into lane 5, and gLIG solution into lane 6, all of this are with the same process as for the DNA Ladder (M). Last is the conduction of gel electrophoresis, it is where the gel was run in a voltage of 130V, and a 40 minutes time span. With all these steps, we now have actual results in the simulation of verifying a recombinant plasmid by gel electrophoresis. MATERIALS Materials Used in Restriction Enzyme Digest ➢ Reagents Chemicals that can be used in this experiment: ● Buffer (2.5XB) Solution ● pKAN-R Plasmid (K) Solution ● pARA Plasmid (A) Solution ● Restriction Enzymes (RE) ● Distilled Water (dH₂0) ➢ Micropipetting Equipment Used to measure small volumes ● P2 Pipette ● P20 Pipette ● P2 Tip Box ● P20 Tip Box ➢ Empty Tubes Used to hold solution ● A+ Tube ● A- Tube ● K+ Tube ● K- Tube ➢ Other Equipment ● Microcentrifuge ● Trash ● Floating Tube Rack ● Freezer ● Water Bath ● Ice Bucket Materials Used in Ligating DNA Fragments ➢ Reagents Chemicals that can be used in this experiment: ● Digested pKAN-R (K+) ● Digested pARA (A+) ● Ligation Buffer (5XB) Solution ● DNA Ligase (LIG) Solution ● Distilled Water (dH₂O) ➢ Micropipetting Equipment Used to measure small volumes ● P2 Pipette ● P20 Pipette ● P2 Tip Box ● P20 Tip Box ➢ Empty Tubes Used to hold solution ● Balancing Tube ➢ Other Equipment ● Microcentrifuge ● Trash ● Floating Tube Rack ● Freezer ● Water Bath ● Ice Bucket ● Vortex Materials Used in Verifying a Recombinant Plasmid by Gel Electrophoresis ➢ Reagents Chemicals that can be used in this experiment: ● pKAN-R (K-) Solution ● pKAN-R (K+) Solution ● pARA (A-) Solution ● pARA (A+) Solution ● Ligated Plasmids (LIG) Solution ● Loading Dye (LD) Solution ● Distilled Water (dH₂0) ● DNA Ladder (M) Solution ● Balancing Tube ● 1 X Sodium Borate Buffer ➢ Micropipetting Equipment Used to measure small volumes ● P2 Pipette ● P20 Pipette ● P2 Tip Box ● P20 Tip Box ➢ Empty Tubes Used to hold solution ● gK- Tube ● gK+ Tube ● gA- Tube ● gA+ Tube ● gLIG Tube ➢ Micropipetting Equipment Used to sort molecules by size and charge ● Agarose Gel ● Gel Electro-phoresis Box ● Power Supply ● Power Supply Leads ➢ Other Equipment ● Microcentrifuge ● Trash RESULTS Figure 1 (Results of gK - ) Predicted Results Actual Results Ideal Results Figure 2 (Results of gK + ) Predicted Results Actual Results Ideal Results Figure 3 (Results of gA - ) Predicted Results Actual Results Figure 4 (Results of gA + ) Ideal Results Predicted Results Actual Results Ideal Results Figure 5 (Results of gLIG) Predicted Results Actual Results Ideal Results In the simulation of verifying a recombinant plasmid by gel electrophoresis, we have come up with the predicted and actual results. The predicted results were made before the simulation process. As we can see, the gK- in figure 1 and the gA- in figure 3 has the same results. It shows that their band pattern in predicted results is different from the actual results, while the band pattern of ideal results is identical to its actual results. Then, for the gK+ or figure 2 and gA+ or figure 4 we can see that the predicted result is just the same as the first two figures (fig. 1 and 3) which is also different from the pattern of both the actual and ideal results. However, the ideal and actual results of figure 4 show that the gel has run to the end because the plasmid in figure 4 travels the most distance from the well. On the other hand, the results of the gLIG or figure 5 were different from each other. It is because our group assumed that the plasmids in the LIG tube are all behaving like DNA fragments and did not consider the different configurations of the recombinant plasmids. In this simulation results, you can see that all the predictions in each figure are different from the actual results. It is because of the ligated plasmids that could occur in configurations that make it difficult to estimate their size. CONCLUSION In conclusion, it is important to verify restriction digest and ligation procedures worked in a recombinant plasmid. It is necessary to verify a recombinant plasmid because multiple plasmids and potential errors are possible to occur, and we wanted to make sure that the expected plasmid was successfully generated. Using gel electrophoresis, we are able to compare the sizes of the restriction digest fragments and ligation product to ensure that the desired recombinant plasmid was obtained. Also, gel electrophoresis allows the separation of genetic material into distinct bands according to molecular size by applying an electric current to an agarose gel. Since the genetic molecules are not visible with the naked eye, DNA loading dye is added to each sample to visualize how fast the gel is running. This keeps the sample from migrating all the way through the gel and getting lost in the surrounding buffer that conducts the current. In addition, gel electrophoresis is used to determine whether the gene of interest and all required components have been successfully included in the recombinant plasmid. Moreover, upon knowing the sizes of the DNA fragments used to create the desired recombinant plasmid, we can visually confirm that we have the desired plasmid by comparing its size to a DNA ladder. A ladder is a mixture of DNA fragments of known size that can be used to estimate the size in base pairs of any plasmid or fragment of unknown length. Apart from this, DNA ligase joins complementary DNA fragments together to produce a recombinant plasmid.