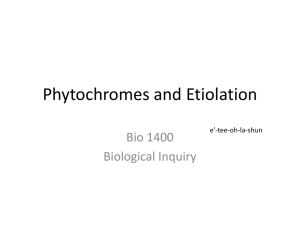
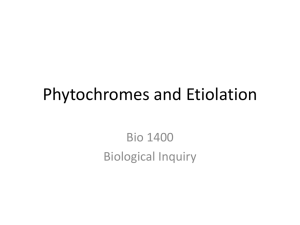
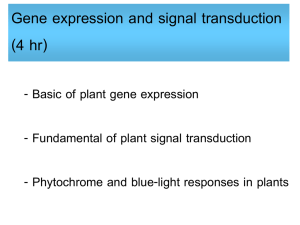
Available online at www.sciencedirect.com ScienceDirect The family of phytochrome-like photoreceptors: diverse, complex and multi-colored, but very useful Katrin Anders and Lars-Oliver Essen Bilin-dependent GAF domain photoreceptors cover the whole spectrum of light with their absorbance properties. They can be divided into three groups according to the domain architecture of their photosensory module. Group I and Group II harbor phytochromes with PAS-GAF-PHY and GAF-PHY domain architecture, respectively. Group III consists of stand-alone GAF domain photoreceptors, the cyanobacteriochromes. Crystal structures of all three groups are now available to shed light on possible downstream signaling pathways. Structures of Group I and III photoreceptors in both states display changes in the secondary structures during photoconversion. The knowledge about the photoconversion in phytochromes and CBCRs make them promising targets for applications in life science and synthetic biology. Address Department of Chemistry, Philipps-University, Hans-Meerwein-Str. 4, D-35032 Marburg, Germany Corresponding author: Essen, Lars-Oliver (essen@chemie.uni-marburg.de) Current Opinion in Structural Biology 2015, 35:7–16 chromophores into three subfamilies dependent on their common domain architecture (Figure 1a). Group I includes canonical phytochromes whose sensory region adopts a PAS-GAF-PHY domain architecture. Firstly discovered in plants this domain organization is characteristic of bacterial, algal and fungal phytochromes as well (Figure 1a). One consequence of their complex topology is a figure-of-eight knot structure, where the variable Nterminus is threaded through an elongated loop of the GAF domain [3,4] (Figure 2a). Cyanobacteria harbor diverse phytochrome-like photoreceptors of Groups II and III (Figure 1a). Cyanobacterial phytochromes of Group II (Cph2) lack only the N-terminal PAS domain, whereas Group III, the cyanobacteriochromes (CBCRs), form a diverse subfamily of bilin-binding stand-alone GAF domains. Together with their effector domains most of these phytochromes apparently exist as dimers. Accordingly, the crystal structures of their photosensory modules are often found as parallel or antiparallel dimers. Interestingly, the two photostates of Group III members can cover any region of light from ultraviolet (UV) to near infrared radiation (NIR). This review comes from a themed issue on Catalysis and regulation Edited by Judith P Klinman and Amy C Rozenzweig http://dx.doi.org/10.1016/j.sbi.2015.07.005 0959-440/# 2015 Elsevier Ltd. All rights reserved. Introduction Light perception allows life to adapt to changing illumination, for example, for controlling morphogenesis, photosynthesis or avoidance of harmful radiation. Phytochromes utilize bilins as chromophore and occur in most kingdoms of life, for example, algae, plants and cyanobacteria, as well as in non-photosynthetic organisms such as bacteria and fungi, but interestingly not in animals and archaea [1]. Their chromophore senses two different light qualities by adopting two stable, photoconvertible isomers: In plant, fungal and many bacterial phytochromes the Pr state absorbs red light to convert to the Pfr state, whereas the latter converts back by far-red light absorption. Furthermore, conversion can proceed independently of light as dark reversion [1]. Rockwell et al. [2] classified GAF domain photoreceptors with bilin www.sciencedirect.com As linear tetrapyrroles, bilins form long-lived excited states by their highly conjugated ring system [5] without being toxic to the cell while allowing enhancement and fine-tuning of their absorption by the protein environment. Bacteriophytochromes (BphP) and fungal phytochromes (Fph) link biliverdin IXa (BV), a degradation product of heme, autocatalytically to an N-terminal cysteine residue as chromophore. Bilin reductases catalyze BV reduction to the other phytobilins (Figure 1b), phytochromobilin (PFB) and phycocyanobilin (PCB) from plant and cyanobacterial phytochromes that are attached to a conserved cysteine in the GAF domain [3,4,6]. The photocycle of these bilin chromophores depends on the Z ! E and E ! Z photoisomerization of their C15 = C16 double bond (Figure 1b). BV-binding phytochromes absorb NIR by their 15E-state and are thus the most red-shifted GAF-containing photoreceptors. So far, all bathyphytochromes (bathy-BphPs) that form Pfr as ground state belong to this group of BV-binding phytochromes. Compared to bathy-BphPs PFB-binding plant phytochromes are blue-shifted for their 15Z-states and 15E-states (Figure 1c). This is further pronounced in PCB-binding phytochromes of Groups I and II with the latter displaying the most blue-shifted absorbance maxima, especially in the Pr state [7]. In contrast, CBCRs undergo very diverse photocycles [8–15] with currently four major types: Two types have opposite photocycles Current Opinion in Structural Biology 2015, 35:7–16 8 Catalysis and regulation Figure 1 Group II (a) PAS (Cph2-like) GAF PHY HKR Group I (canonical) PhyA PhyB R Cys R Cys N SynCph1 MCP HK GAF PAS PHY TePixJ R Cys MCP AnPixJ N HK RpBphP2 DrBphP NcFph1 SynCph2 HK RR EAL (b) O O O BV O (15Z) C B NH HN H N 15 16 H N D O A Cys O Group III (CBCR) O O O PCB B O R Cys R Cys R C NH HN H N 15 16 H N D O Cys R A O (c) Cys 760 R O O O PΦB B C NH HN H N 15 16 O H N 15 16 D N H O O O Cys C B NH O D HN H N A 15 16 A O Cys O PVB O O Cys H N D O Absorbance maximum 15E-state (nm) BV Pfr (15E) Phytochromes 720 Pg/Pr CBCRs 680 640 Insert-Cys and DxCF CBCRs 600 560 520 Pr/Pg CBCRs 480 400 440 480 520 560 600 640 680 720 Absorbance maximum 15Z-state (nm) Current Opinion in Structural Biology Domain architecture, chromophores and spectral diversity of members of the phytochrome family. (a) Sequence-similarity network of phytochrome-like GAF domains (INTERPRO entry 16032; edges correspond to pair-wise BLAST E-values of <1075; each node to sequences of >70% sequence identity) as calculated by the EFI-enzyme similarity web tool (efi.igb.illinois.edu/efi-est). Group I phytochromes are clustered in plant (light-red), bacterial (dark-red) and fungal (brown) subfamilies. PAS and PHY domains are depicted in red and green, respectively. Blue stars indicate the bilin chromophore and the location of the cysteine as chromophore attachment site that is either part of the GAF domain (light red) or the N-terminus (GAF domain: dark red). Red lines depict the knot regions, dark-green loops the tongue-region of the PHY domain. Effector domains are abbreviated by R; CBCRs (gray) may also have N-terminal domains. Domain organizations of Group I, II (orange) and III (gray, black) members including the effector domains are shown as ellipses and rectangles (PAS: red; effector domains: white, gray or black; HKR domain: histidine kinase related domain; Syn: Synechocystis sp. PCC 6803; Nc: Neurospora crassa; An: Anabaena sp. PCC 7120; Te: Thermosynechococcus elongates; Rp: Rhodopseudomonas palustris). Current Opinion in Structural Biology 2015, 35:7–16 www.sciencedirect.com Phytochromes: diverse, complex and multi-colored Anders and Essen 9 with green/red (G/R) CBCRs adopting a green-absorbing (15Z) ground state and a red-absorbing (15E) photoproduct [16], whereas red/green (R/G) CBCRs convert from a redabsorbing (15Z) state to a green-absorbing (15E) photoproduct [8,15,17,18]. The latter shows that the 15E-isomer is not necessarily the long-wavelength absorbing state. The insert-Cys and DxCF-types of CBCRs employ a second cysteine residue for additional chromophore attachment [14,19]. Their 15Z ground state absorbs at shorter wavelengths from near-UV to blue, whereas the 15E-state absorbs at longer wavelengths, that is, from blue to orange. All CBCRs attach initially PCB as chromophore, but DXCF-type CBCRs autocatalytically isomerize PCB into PVB, thus shifting the absorbance of the 15E-state between teal and orange [12,13,20]. Interestingly, algae harbor Group I phytochromes, which clearly deviate from Pr/ Pfr-phytochromes of plant and fungal origin by exerting a similar capability of spectral tuning as the CBCRs [21,22]. The structural repertoire of phytochrome-like photoreceptors Group I phytochromes show common features for binding of BV, PFB or PVB in their GAF domain (Figure 2a). Besides being involved in knot formation the N-terminus clings to the chromophore-binding site and additionally shields the chromophore from solvent access. The GAF domain is connected via a long helical spine to the PHY domain. The latter protrudes a tongue-like region that reaches towards the GAF domain and covers the chromophore-binding pocket. The Group II phytochrome structure [23] closely resembles structurally Group I, but lacks the N-terminal PAS domain and therefore the knot in the structure. Both, in Group I and II structures the tongue region shows common features depending on the photochemical state of the protein. In the Pr-state the stem of the tongue region is built by a two-stranded b-hairpin-like structure. The length of the b-strands and the region at the tip of the tongue vary but the positions of the conserved WG/AG, PRxSF and WxE motifs are invariant [3,23] (Figure 2c, left). The Pfr structures of bathy-BphPs show different features [4,24] (Figure 2c, right). Here, loop and a-helical sections build the stem region. The tip of the tongue does not vary in length but in the orientation of the loop region. The WG/AG, PRxSF and WxE motifs are conserved in their positions but not as strictly as in the Pr structures. The photosensory GAF domain of CBCRs is structurally highly related to Groups I and II apart from a truncated loop region that is part of the knot structure in Group I phytochromes and forms an additional b-strand in knotless Group II GAF domains (Figure 2b, red box). Another difference is the a-helix facing the chromophore in nonBphPs that is distinctly oriented in Group I, II and III photoreceptors. In addition, the loop region above the chromophore-binding site differs (Figure 2b, inlet). Here, group I and II phytochromes form a short helix that includes the conserved DIP motif. The aspartate of this motif is part of a salt bridge in Group I and II phytochromes that connects the tongue region of the PHY domain with the GAF domain, the course of the following loop/b-strand region is similar in both groups. In CBCRs the DIP motif helix is missing, the subsequent loop regions are more shifted towards the PCB-binding a-helix thus narrowing the cleft that leads to the bilin-binding site, which is otherwise blocked by the missing PHY domain. In the TePixJ (15E) structure [25] (Figure 2b inlet, blue) this loop region is shifted to an extent that it nearly closes the cleft. Photoconversion and down-stream signaling in phytochromes and CBCRs Structural changes upon photoconversion have been intensely studied to elucidate intramolecular pathways for downstream signaling. 15Z-structures and 15E-structures are now available both for phytochromes and CBCRs. Structures of the Group I phytochrome DrBphP from Deinococcus radiodurans (Figure 3a–c) were solved in its Pr (15Z) and Pfr (15E) state [26] and confirmed an earlier Trp-switch model [23]. Here, Z ! E isomerization by red light triggers structural changes, where an aspartatearginine salt bridge is broken with an accompanying swap of tryptophans from the WG/AG and WxE motifs within the GAF/tongue interface. The GAF-PHY domain linking helical spine is straightened during photoconversion (Figure 3b), whereas the tongue undergoes further refolding from a b-hairpin-like to an a-helical structure, a b/atransition inferred before from differences between Pr and bathy-BphP Pfr tongue structures (Figure 2c) [23,27]. The PRxSF motif alters its conformation such that new interactions are formed between the tongue and the GAF domain [23] (Figure 4a). These changes also affect the C-terminal a-helix that connects the photosensory with the effector module [26]. Single particle electron microscopy (SPEM) of DrBphP full length dimers in the Pr and Pfr conformation indicate a quaternary structure for both states [28], which corresponds to a parallel head-to-head and intimately twisted arrangement ( Figure 1 Legend Continued ) The central part of the panel comprises isolated clusters, which are related to other Group I-III phytochromes with E values of >1075. (b) Chromophores of phytochrome-like photoreceptors; BV: biliverdin; PCB: phycocyanobilin, PFB: phytochromobilin; PVB: phycoviolobilin in the Pr (15Z) conformation. Differences relative to BV as well as the Pfr (15E) conformation of the C15-C16 double bond are highlighted in red. (c) Absorbance maxima of the 15Z-states and 15E-states of photosensory modules from photoreceptors of Groups I-III. Black squares represent Group I phytochromes; cyan colored triangles show bathy-BphPs with a Pfr ground state that also belong to this group. Red dots indicate members of Group II, green triangles members of Group III, the CBCRs. The black line is a diagonal in the wavelength scale indicating either red shift (above) or blue shift (below) upon Z ! E photoisomerisation. Black frames show the assignment of phytochrome-like proteins to subfamilies, red circles unusual CBCRs or phytochromes. www.sciencedirect.com Current Opinion in Structural Biology 2015, 35:7–16 10 Catalysis and regulation Figure 2 (a) Group I tongue (b) AtPhyB C GAF PAS PHY N SynCph1 AnPixJ TePixJ_15E 90º C N PaBphP Group SynCph1 I SynCph2 II TePixJ_15Z III C Group II SynCph2 C N Group III: CBCRs (c) Tongue SynCph2 Tongue SynCph1 AnPixJ GAF S W Tongue PaBphP Tongue RpBphP1 W F R x C W GAF P R F W P x N S PCB TePixJ_15E PAS N Pr BV Pfr C Current Opinion in Structural Biology Phytochrome and CBCR structures. (a) Structure of the photosensory modules of Group I–III photoreceptors. PAS, GAF and PHY domains are shown in red, orange/violet/gray and green, respectively. The chromophore is displayed in cyan. PDB-codes: AtPhyB: 4OUR, SynCph1: 2VEA, PaBphP: 3NHQ, SynCph2: 4BWI, AnPixJ: 3W2Z, TePixJ (15Z): 4GLQ, 4FOF, TePixJ (15E): 3VV4. (b) Superimposition of the SynCph1 (Group I), SynCph2 (Group II) and TePixJ (15Z) (Group III) GAF domains; the inlet and boxes illustrate differences between the GAF domains. The inset displays a magnification of the chromophore-binding site regarded from the top. In addition, also AnPixJ and TePixJ (15E) are shown in this inlet. The lower red box shows the knot-forming loop region in Group I phytochromes at the end of the b-sheet in the GAF domain. (c) Comparison of the tongue region in the Pr conformation of SynCph1 and SynCph2 and the Pfr conformation of PaBphP and RpBphP1 (PDB code: 4GW9). Conserved amino acids in the PRxSF motif as well as the tryptophan residues of the WG/AG and WxE motif are shown in stick representation. Ca-atoms of the b-sheets in the tongue of the Pr state are indicated by red and blue dots so that their position in the Pfr state can be located. Current Opinion in Structural Biology 2015, 35:7–16 www.sciencedirect.com Phytochromes: diverse, complex and multi-colored Anders and Essen 11 Figure 3 (a) Group I DrBphP Tongue Pr GAF N GAF S x F Y P F x W R N R Y P S BV C Tongue Pfr BV C GAF: Pr GAF: Pfr PAS: Pr PAS: Pfr (b) Pr PAS PAS PHY: Pr PHY: Pfr Pr (c) Pfr Pfr (d) Pr Pfr PHY PHY 9Å PAS / GAF CBCR HK 35º 124 Å PAS / GAF 85 Å (e) HK HK HK 143 Å Pfr’ HK HK 90 Å 35º PHY 5Å 124 Å PAS / GAF 86 Å PVB 15Z PVB 15E PVB 15Z Cys A TePixJ 15Z TePixJ 15E D C B Cys Cys PVB 15E A D N C B C C Current Opinion in Structural Biology Comparison of the 15Z and 15E structures in Group I phytochrome and Group III CBCR structures. (a) Overlay of the Pr and Pfr structures of DrBphP (PDB codes: 4O0P, 4O01). PAS; GAF, PHY domains and BV are displayed in red, orange, green and cyan, respectively. The Pfr structure is represented in pale colors. The inlets show the tongue region of DrBphP in the Pr (left) and Pfr (right) photostate. The amino acids of the PRxSF motif as well as the tryptophan and tyrosine residue of the WG/AG and W/YxE motifs are presented. (b,c) Illustration of the PHY domain movement upon Pfr formation in DrBphP, regarded from the top looking on the tongue region (b) or in the dimer with side view (c). (d) Single particle electron microscopy maps of the DrBphP dimer in the Pr, as well as in the Pfr and P0fr conformation [28]; surface-rendered front and side views with the included Pr crystal structure of DrBphP (PDB code: 4Q0J) docked as rigid body. In this structure PAS, GAF and PHY domains are shown in blue, green and orange, respectively, and the other monomer is displayed in gray. The PHY domain is rotated by 358 clockwise and moved outside by 9 Å in Pfr and by 5 Å in P0fr . (e) Comparison of the 15Z-structures and 15E-structures of TePixJ. Left: overall structure; middle: view on the chromophore binding pocket from the top; right side: the PVB chromophore of TePixJ. www.sciencedirect.com Current Opinion in Structural Biology 2015, 35:7–16 12 Catalysis and regulation Figure 4 (a) (b) GAF1 DrBphP: Pr DrBphP: Pfr GAF Pr TePixJ 15Z TePixJ 15E 2 C D F G G R W E P Wx S x C ole ind C C ~45º (c) R R GAF1 Pfr W P F W x R X N N 2 S GAF D E G G (d) EAL EAL * GGDEF GGDE F* GGDEF* GGDEF* EAL EAL HK P HK HK HK P Current Opinion in Structural Biology Signal transduction pathways in phytochromes and CBCRs. (a) Model for conformational changes within the tongue region during photoconversion [23]. Upon red-light illumination the aspartate–arginine salt bridge between the GAF1 domain and the tongue (inlet) is broken. The tip of the tongue refolds and induces a disordering in the b-sheet stalk region while the tryptophan motifs are switched and the new aspartate–serine hydrogen bond (inlet) is formed. (b) C-terminal a-helices of the photosensory modules from DrBphP (left) and TePixJ (right). In DrBphP the helix is shifted from the Pr to the Pfr state, in TePixJ only a minor shift takes place but the helix is rotated about 458. (c,d) Different mechanisms for downstream signaling by phytochrome-like photoreceptors. PAS, GAF and PHY domains are shown in red, orange and green, respectively. The chromophore is displayed as a blue star. Arrows indicate illumination with red or far red light to achieve the Pfr or Pr state, respectively. The blue ellipse shows a possible binding partner, the gray flash indicates the active state. Red circles indicate substrate molecules. (c) Intermolecular interaction of the phytochrome with a possible binding partner in the Pfr state. (d) Intramolecular interactions: the structural changes upon photoconversion to Pfr lead to a rearrangement of the HK domains, allowing the trans-phosphorylation (left). Structural rearrangements lead to a better access to the active site of the effector domain thus activating the domain (right). Current Opinion in Structural Biology 2015, 35:7–16 www.sciencedirect.com Phytochromes: diverse, complex and multi-colored Anders and Essen 13 of the two monomers. For Pr EM maps reveal a central hole and a C-terminal bulky density reflecting the paired HKs (Figure 3d). The Pr crystal structure of DrBphP can be docked into the map, although the helical spine is placed in a central hole of the map without being covered by density. Photoactivation apparently generates two populations of particles, Pfr and P0fr (Figure 3d, middle, right), with an increased diameter of the central hole and altered arrangements of the PHY domains. Interestingly, upon docking the Pfr DrBphP crystal structure in the SPEM map, the PHY domain has to be rotated and displaced to fit the Pfr and P0fr conformations [28]. The change of the chromophore itself upon photoconversion is limited to the Z ! E double bond isomerization between C15 and C16 and minor relaxations of the molecule that can be also observed in cryo-trapped intermediate states [29]. Evidences for a possible A-ring instead of a D-ring rotation in Group II phytochromes upon photoconversion [30] could be disproved [31] and derived apparently from the high solvent accessibility of the PCB chromophore along rings A and B [23]. Recombinant CBCRs usually contain a mixture of PCB and PVB [20]. Accordingly, the structure of the B/Gabsorbing DxCF-type CBCR TePixJ from Thermosynechococcus elongatus was analyzed in its PVB-bound form as Pb (C10-Zs/C15-Za) [25] and Pg (C10-Zs/C15-Ea) state [32], respectively, as well as in the PCB-bound form as Pb state [25]. The Pb states of TePixJ exhibit a second thioether linkage between the C10 carbon of the chromophore and the cysteine from the conserved DxCF motif [25]. This causes a rubinoid bilin species with shortened p-system, whereas in the Pg state this thioether linkage is broken [32]. Conformational changes occur mainly in regions of the chromophorebinding pocket (Figure 3e), where in Group I and II phytochromes the tongue shields the chromophore. During Z ! E photoconversion, the terminal strand of the b-sheet diminishes solvent access to the chromophore by converting into a flexible region and a 310-helix that moves towards the PVB-binding a-helix. Like in phytochromes [24,33] the C-terminal a-helix of the GAF domain in AnPixJ and TePixJ continues as helix into the downstream effector domain [32]. As these helical spines apparently transmit signals over long distances [4] this feature is probably not only crucial for signal transduction by TePixJ [34], but generally for all GAF-containing photoreceptors. Light-triggered release of the terminal b-strand in TePixJ may be hence crucial for downstream signaling, as its N-terminal and C-terminal a-helices are coupled to the b-strands [34]. Additionally, an axial rotation of the C-terminal helix by 458 during 15Z ! 15E photoconversion (Figure 4b) transmits to an altered orientations of the flanking HAMP domains [34], which are known to act to as rigid transmitters by rotation of the helix bundles in their dimerization interface [35]. www.sciencedirect.com Synthetic biology applications for phytochromes Near-infrared is attractive for applications in life sciences, because autofluorescence, scattering and absorption by cellular components is mostly negligible thus allowing photocontrol in thick tissues and even live animals [36]. As Group I phytochromes have fluorescence and action spectra in this wavelength range, they are promising templates for the engineering of biomarkers, biosensors and optogenetic switches. To engineer NIR-fluorescent phytochromes the stability of their Pr conformation is optimized by (a) hindrance of the C15 = C16 double bond isomerization after photoexcitation, (b) destabilization of the chromophore formed after photoisomerization to an early intermediate called Lumi-R, and (c), blockage of its deprotonation transiently occurring during the further Pr ! Pfr photoconversion [36]. Removal of the PHY domain also fosters fluorescence properties as in Wi-Phy, a monomeric PAS-GAF variant of DrBphP, that harbors mutations in the dimerization interface as well as for the DIP motif’s aspartate and Y263 in the D-ring environment [36]. At least one WiPhy variant is applicable as NIR-fluorescent biomarker in mammalian cells and mice, where it incorporates endogenous BV [37]. Resonance-Raman and molecular dynamics studies on iRFP, a biomarker based on RpBphP2 from Rhodopseudomonas palustris, indicate that the chromophore is distorted relative to wild type by an increased tilt angle between rings C and D and a loss of hydrogen bond interactions to the ring D carbonyl. Here, subsequent loss of water molecules in the binding pocket [38] may cause rigidification of the chromophore-binding domain and thus a decrease of non-radiative decay processes. Likewise, the IFP1.4 biomarker derived from DrBphP shows tight hydrophobic packing around ring D [39] and very long life times for excited Pr states. Nevertheless, current phytochrome-based NIR-biomarkers still have a restricted potential due to quantum yields of <10%. Phytochromes are proposed as fluorescent sensors for redox potentials, metal ions or protein-protein interactions by exploiting their modular multidomain organization and potential for spectral fine-tuning by altering their chromophore or 3D-structure. For example, a BphPbased biosensor for mercury uses the high affinity of Hg2+ ions to cysteines for competing with BV-attachment [40]. To detect protein-protein interactions the PAS and GAF domains of RpBphP2 are employed as iSplit biosensors. Here, these domains are parts of separate polypeptide chains, each fused to a possible interaction partner, respectively. Upon association the PAS and GAF domains assemble so that BV gets covalently attached and the iSplit phytochrome starts to fluoresce. This split NIR-biosensor functions in human cell lines and whole mice [41]. The third direction of phytochrome engineering is optogenetics with currently three types of Current Opinion in Structural Biology 2015, 35:7–16 14 Catalysis and regulation applications: The photosensory module of SynCph1 was fused to the HK region of EnvZ to control HK activity by red light for repressing the expression of OmpR-dependent genes [42,43]. Alternatively, the light-regulated PhyB-PIF3 interaction can control gene expression, e.g. by splitting the DNA-binding and transactivation domains of GAL4 [44], or co-localize target proteins to subcellular components [45–47]. Finally, a DrBphP-phosphodiesterase fusion steers cyclic nucleotide levels by light in vivo [48]. Conclusions Overall, the mechanistic understanding of phytochromelike photoreceptors faces two challenges: Firstly, how do we use structural and spectroscopic data for a concise understanding of color tuning? Given the difficulty to rationalize color-tuning in microbial and animal rhodopsins, which depend on the ‘simpler’ chromophore retinal, it is not surprising that theoretical approaches such as full-scale quantum chemical and QM/MM calculations on complex bilin chromophores in their protein environment are still in their infancy [49–51]. Here, progress is not only impeded by radiation effects on the chromophore during structural analyses [23,52], but also by intrinsic conformational heterogeneity of the Pr state and [53–56] and to some extent also the Pfr state [57]. The second issue is the chosen mode of downstream signaling, which apparently differs in the family of phytochrome-like photoreceptors (Figure 4c and d). So far, we are missing structural views how plant phytochromes form light-dependent complexes with the plethora of known phytochrome-interacting factors such as COP1, PIFs or FHY1. In terms of bacterial phytochromes we can at least extrapolate the activation/inactivation of effector domains by extrapolating from other photosensory systems [58]. One pressing issue for synthetic phytochromes to be applicable in optogenetics is chromophore supply, as animals provide only BV from heme breakdown. Accordingly, PCB production requires at least two additional transgenes [59] and makes BphPs to more attractive templates. Another challenge is the complexity of the architecture of Group I and II phytochromes. Here, CBCRs indeed excel by high fluorescence, e.g. GAF3 from the RGS protein from Synechocystis sp. displays a twofold higher fluorescence quantum yield than SynCph2 (1-2) [7,60]. Furthermore, CBCRs are now known, which do not only incorporate PCB as chromophore, but also the better bioavailable BV [17]. In this context, the small size and high spectral diversity of CBCRs calls for their future engineering and application. Conflict of interest statement No conflict of interest. Current Opinion in Structural Biology 2015, 35:7–16 Acknowledgements The authors are grateful for funding by the Deutsche Forschungsgemeinschaft (ES152/9, ES152/10) and the LOEWE-Center for Synthetic Microbiology. References and recommended reading Papers of particular interest, published within the period of review, have been highlighted as: of special interest of outstanding interest 1. Rockwell NC, Su YS, Lagarias JC: Phytochrome structure and signaling mechanisms. Annu Rev Plant Biol 2006, 57:837-858. 2. Rockwell NC, Lagarias JC: A brief history of phytochromes. ChemPhysChem 2010, 11:1172-1180. 3. Essen LO, Mailliet J, Hughes J: The structure of a complete phytochrome sensory module in the Pr ground state. Proc Natl Acad Sci U S A 2008, 105:14709-14714. 4. Yang X, Kuk J, Moffat K: Crystal structure of Pseudomonas aeruginosa bacteriophytochrome: photoconversion and signal transduction. Proc Natl Acad Sci U S A 2008, 105:14715-14720. 5. Scheer H, Zhao KH: Biliprotein maturation: the chromophore attachment. Mol Microbiol 2008, 68:263-276. 6. Burgie ES, Bussell AN, Walker JM, Dubiel K, Vierstra RD: Crystal structure of the photosensing module from a red/far-red lightabsorbing plant phytochrome. Proc Natl Acad Sci U S A 2014, 111:10179-10184. The crystal structure of the ‘most important’ photoreceptor on Earth, phytochrome B, a major determinant of crop yields, corroborates the role of conserved aromatic residues in the tongue region for stabilizing the distinct tongue/GAF packings in Pr and Pfr states. 7. Anders K, von Stetten D, Mailliet J, Kiontke S, Sineshchekov VA, Hildebrandt P, Hughes J, Essen L-O: Spectroscopic and photochemical characterization of the red-light sensitive photosensory module of Cph2 from Synechocystis PCC 6803. Photochem Photobiol 2011, 87:160-173. 8. Narikawa R, Enomoto G, Ni Ni W, Fushimi K, Ikeuchi M: A new type of dual-Cys cyanobacteriochrome GAF domain found in cyanobacterium Acaryochloris marina, which has an unusual red/blue reversible photoconversion cycle. Biochemistry 2014, 53:5051-5059. AM1_1186 is related to R/G CBCRs, but employs unlike these a second cysteine for reversible, light-dependent bond formation to the PCB chromophore. Its red/blue photochromicity is currently record holder with a triggered wavelength shift of 225 nm. 9. Lim S, Rockwell NC, Martin SS, Dallas JL, Lagarias JC, Ames JB: Photoconversion changes bilin chromophore conjugation and protein secondary structure in the violet/orange cyanobacteriochrome NpF2163g3. Photochem Photobiol Sci 2014, 13:951-962. NMR shift analyses show that the C-terminal helix of the first NpF2164g3 CBCR domain becomes ordered, whereas the insert-Cys loop destabilized during photoconversion. 10. Hauck AFE, Hardman SJO, Kutta RJ, Greetham GM, Heyes DJ, Scrutton NS: The photoinitiated reaction pathway of full-length cyanobacteriochrome Tlr0924 monitored over 12 orders of magnitude. J Biol Chem 2014, 289:17747-17757. This DxCF-type CBCR forms 15E-PVB’/15E-PCB’ intermediates within 10 ps, but the subsequent conformational changes of the protein matrix to the final photoproducts require seconds. 11. Velazquez Escobar F, Utesch T, Narikawa R, Ikeuchi M, Mroginski MA, Gärtner W, Hildebrandt P: Photoconversion mechanism of the second GAF domain of cyanobacteriochrome AnPixJ and the cofactor structure of its green-absorbing state. Biochemistry 2013, 52:4871-4880. 12. Rockwell NC, Martin SS, Gulevich AG, Lagarias JC: Phycoviolobilin formation and spectral tuning in the DXCF cyanobacteriochrome subfamily. Biochemistry 2012, 51:1449-1463. www.sciencedirect.com Phytochromes: diverse, complex and multi-colored Anders and Essen 15 13. Enomoto G, Hirose Y, Narikawa R, Ikeuchi M: Thiol-based photocycle of the blue and teal light-sensing cyanobacteriochrome Tlr1999. Biochemistry 2012, 51:3050-3058. 14. Rockwell NC, Martin SS, Feoktistova K, Lagarias JC: Diverse two-cysteine photocycles in phytochromes and cyanobacteriochromes. Proc Natl Acad Sci U S A 2011, 108:11854-11859. 15. Narikawa R, Fukushima Y, Ishizuka T, Itoh S, Ikeuchi M: A novel photoactive GAF domain of cyanobacteriochrome AnPixJ that shows reversible green/red photoconversion. J Mol Biol 2008, 380:844-855. 16. Hirose Y, Shimada T, Narikawa R, Katayama M, Ikeuchi M: Cyanobacteriochrome CcaS is the green light receptor that induces the expression of phycobilisome linker protein. Proc Natl Acad Sci U S A 2008, 105:9528-9533. 17. Narikawa R, Nakajima T, Aono Y, Fushimi K, Enomoto G, Ni Ni W, Itoh S, Sato M, Ikeuchi M: A biliverdin-binding cyanobacteriochrome from the chlorophyll d-bearing cyanobacterium Acaryochloris marina. Sci Rep 2015, 5:7950. The R/G CBCR AM1_1557 is capable to incorporate BV instead of PCB as chromophore and is thus highly interesting for optogenetic applications. 18. Gottlieb SM, Kim PW, Chang C-W, Hanke SJ, Hayer RJ, Rockwell NC, Martin SS, Lagarias JC, Larsen DS: Conservation and diversity in the primary forward photodynamics of red/green cyanobacteriochromes. Biochemistry 2015, 54:1028-1042. 19. Rockwell NC, Martin SS, Lagarias JC: Identification of DXCF cyanobacteriochrome lineages with predictable photocycles. Photochem Photobiol Sci 2015:20. 20. Ishizuka T, Kamiya A, Suzuki H, Narikawa R, Noguchi T, Kohchi T, Inomata K, Ikeuchi M: The cyanobacteriochrome TePixJ, isomerizes its own chromophore by converting phycocyanobilin to phycoviolobilin. Biochemistry 2011, 50:953-961. 21. Rockwell NC, Duanmu D, Martin SS, Bachy C, Price DC, Bhattacharya D, Worden AZ, Lagarias JC: Eukaryotic algal phytochromes span the visible spectrum. Proc Natl Acad Sci U S A 2014, 111:3871-3876. By analyzing the available genomic function of algal species and therefrom derived recombinant phytochromes this report shows that the two photostates of algal Group I phytochromes cover the whole visible spectrum like cyanobacterial CBCRs. 22. Duanmu D, Bachy C, Sudek S, Wong C-H, Jiménez V, Rockwell NC, Martin SS, Ngan CY, Reistetter EN, van Baren MJ et al.: Marine algae and land plants share conserved phytochrome signaling systems. Proc Natl Acad Sci U S A 2014, 111:15827-15832. Evolutionary, the classical Pr/Pfr-switching phytochromes of land plants are derived from algal ancestors with their much wider degree of spectral tuning. 23. Anders K, Daminelli-Widany G, Mroginski MA, von Stetten D, Essen L-O: Structure of the cyanobacterial phytochrome 2 photosensor implies a tryptophan switch for phytochrome signaling. J Biol Chem 2013, 288:35714-35725. The first crystal structure of a Group II phytochrome allowed to predict the conformational switching of the tongue regions upon photoconversion. Combinig crystallography with SAXS and MD simulations compelling evidence for large-scale changes of the domain and tongue organisation upon photoconversion of DrBphP is shown. Interestingly, the Pfr state crystal structure with its splayed PHY domains is not fully representative for a solution-state conformation. 27. Anders K, Gutt A, Gaertner W, Essen L-O: Phototransformation of the red-light sensor cyanobacterial phytochrome 2 from Synechocystis sp. depends on its tongue motifs. J Biol Chem 2014, 289:25590-25600. Time-resolved spectroscopy shows that tryptophans of the WxE and WGG tongue motifs are crucial for late intermediate formation. This may reflect slow beta/alpha-transition and altered docking of the tongue to the GAF domain during photoconversion. 28. Burgie ES, Wang T, Bussell AN, Walker M, Li H, Vierstra RD: Crystallographic and electron microscopic analyses of a bacterial phytochrome reveal local and global rearrangements during photoconversion. J Biol Chem 2014, 289:24573-24587. Besides showing the structure of the D207A mutant of DrBphP that binds cyclic hemes instead of PCB the EM data point to large scale structural rearrangements of full-length DrBphP during Pr/Pfr photoconversion. 29. Yang X, Ren Z, Kuk J, Moffat K: Temperature-scan cryocrystallography reveals reaction intermediates in bacteriophytochrome. Nature 2011, 479:428-432. 30. Ulijasz AT, Cornilescu G, Cornilescu CC, Zhang J, Rivera M, Markley JL, Vierstra RD: Structural basis for the photoconversion of a phytochrome to the activated Pfr form. Nature 2010, 463:250-254. 31. Song C, Psakis G, Kopycki J, Lang C, Matysik J, Hughes J: The D-ring, not the A-ring, rotates in Synechococcus OS-B phytochrome. J Biol Chem 2014, 289:2552-2562. The authors resolve by solid-state NMR a long-going debate whether Aring instead of D-ring rotation occurs in Group II phytochromes. Interestingly, chemical shift changes can mislead when being correlated to conformational changes of bilin chromophores. 32. Narikawa R, Ishizuka T, Muraki N, Shiba T, Kurisu G, Ikeuchi M: Structures of cyanobacteriochromes from phototaxis regulators AnPixJ and TePixJ reveal general and specific photoconversion mechanism. Proc Natl Acad Sci U S A 2013, 110:918-923. The crystal structures of the R/G CBCR AnPixJ and the B/G CBCR TePixJ reveal the formation of parallel dimers and predict the methine bridge between rings B and C as attachment site for the second cysteine. 33. Li H, Zhang J, Vierstra RD, Li H: Quaternary organization of a phytochrome dimer as revealed by cryoelectron microscopy. Proc Natl Acad Sci U S A 2010, 107:10872-10877. 34. Cornilescu CC, Cornilescu G, Burgie ES, Markley JL, Ulijasz AT, Vierstra RD: Dynamic structural changes underpin photoconversion of a blue/green cyanobacteriochrome between its dark and photoactivated states. J Biol Chem 2014, 289:3055-3065. Solution state structures of the TePixJ CBCR domain in its Pb and Pg photostates show significant light-induced migration and distortion of the PVB chromophore within the binding site. 35. Ferris HU, Dunin-Horkawicz S, Hornig N, Hulko M, Martin J, Schultz JE, Zeth K, Lupas AN, Coles M: Mechanism of regulation of receptor histidine kinases. Structure 2012, 20:56-66. 24. Bellini D, Papiz MZ: Structure of a bacteriophytochrome and light-stimulated protomer swapping with a gene repressor. Structure 2012, 20:1436-1446. Light-dependent dimerization of the photosensory module of this bathyphytochrome with an unusual effector domain causes the dissociation and thus activation of the latter. 36. Auldridge ME, Satyshur KA, Anstrom DM, Forest KT: Structureguided engineering enhances a phytochrome-based infrared fluorescent protein. J Biol Chem 2012, 287:7000-7009. An monomeric photosensory module was engineered on the base of the PAS-GAF bidomain of DrBphP. The Y263F mutations proved to be superior for fluorescence than substitutions of the DIP motif’s aspartate. 25. Burgie ES, Walker JM, Philipps GNJ, Vierstra RD: A photo-labile thioether linkage to phycoviolobilin provides the foundation for the blue/green photocycles in DXCFcyanobacteriochromes. Structure 2013, 21:88-97. Crystal structures of the TePixJ CBCR domain in its Pb and Pg states. 37. Shu X, Royant A, Lin MZ, Aguilera TA, Lev-Ram V, Steinbach PA, Tsien RY: Mammalian expression of infrared fluorescent proteins engineered from a bacterial phytochrome. Science 2009, 324:804-807. 26. Takala H, Björling A, Berntsson O, Lehtivuori H, Niebling S, Hoernke M, Kosheleva I, Henning R, Menzel A, Ihalainen JA et al.: Signal amplification and transduction in phytochrome photosensors. Nature 2014, 509:245-248. www.sciencedirect.com 38. Velazquez Escobar F, Hildebrandt T, Utesch T, Schmitt FJ, Seuffert I, Michael N, Schulz C, Mroginski MA, Friedrich T, Hildebrandt P: Structural parameters controlling the fluorescence properties of phytochromes. Biochemistry 2014, 53:20-29. Current Opinion in Structural Biology 2015, 35:7–16 16 Catalysis and regulation 39. Bhattacharya S, Auldridge ME, Lehtivuori H, Ihalainen JA, Forest KT: Origins of fluorescence in evolved bacteriophytochromes. J Biol Chem 2014, 289:32144-32152. Rigidification of the chromophore-binding site appears to be a key for well-fluorescent phytochrome-based NIR biosensors. 40. Gu Z, Zhao M, Sheng Y, Bentolila LA, Tang Y: Detection of mercury ion by infrared fluorescent protein and its hydrogelbased paper assay. Anal Chem 2011, 83:2324-2329. 41. Filonov GS, Verkhusha VV: A near-infrared BiFC reporter for in vivo imaging of protein-protein interactions. Chem Biol 2013, 20:1078-1086. A split variant of iRFP, iSplit, assembles with BV concentrations >1 mM when getting in touch by its fusion partners. The quantum-chemical calculation of phytochrome spectra by QM/MM methods depends in the visible regime crucially on the chosen functionals. 51. Mroginski MA, Kaminski S, von Stetten D, Ringsdorf S, Gärtner W, Essen LO, Hildebrandt P: Structure of the chromophore binding pocket in the Pr state of plant phytochrome phyA. J Phys Chem B 2011, 115:1220-1231. 52. Li F, Burgie ES, Yu T, Héroux A, Schatz GC, Vierstra RD, Orville AM: X-ray radiation induces deprotonation of the bilin chromophore in crystalline D. radiodurans phytochrome. J Am Chem Soc 2015, 137:2792-2795. 42. Tabor JJ, Salis HM, Simpson ZB, Chevalier AA, Levskaya A, Marcotte EM, Voigt CA, Ellington AD: A synthetic genetic edge detection program. Cell 2009, 137:1272-1281. 53. Yang Y, Linke M, von Haimberger T, Matute R, González L, Schmieder P, Heyne K: Active and silent chromophore isoforms for phytochrome Pr photoisomerization: an alternative evolutionary strategy to optimize photoreaction quantum yields. Struct Dyn 2014, 1:014701. 43. Levskaya A, Chevalier AA, Tabor JJ, Simpson ZB, Lavery LA, Levy M, Davidson EA, Scouras A, Ellington AD, Marcotte EM et al.: Synthetic biology: engineering Escherichia coli to see light. Nature 2005, 438:441-442. 54. Kim PW, Rockwell NC, Martin SS, Lagarias JC, Larsen DS: Dynamic inhomogeneity in the photodynamics of cyanobacterial phytochrome Cph1. Biochemistry 2014, 53:2818-2826. 44. Shimizu-Sato S, Huq E, Tepperman JM, Quail PH: A lightswitchable gene promoter system. Nat Biotechnol 2002, 20:1041-1044. 55. Song C, Essen LO, Gärtner W, Hughes J, Matysik J: Solid-state NMR spectroscopic study of chromophore-protein interactions in the Pr ground state of plant phytochrome A. Mol Plant 2012, 5:698-715. 45. Beyer HM, Juillot S, Herbst K, Samodelov SL, Müller K, Schamel WW, Romer W, Schafer E, Nagy F, Strahle U et al.: Red light-regulated reversible nuclear localization of proteins in mammalian cells and zebrafish. ACS Synth Biol 2015 http:// dx.doi.org/10.1021/acssynbio.5b00004. 46. Yang X, Jost AP, Weiner OD, Tang C: A light-inducible organelletargeting system for dynamically activating and inactivating signaling in budding yeast. Mol Biol Cell 2013, 24:2419-2430. 47. Levskaya A, Weiner OD, Lim WA, Voigt CA: Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 2009, 461:997-1001. 48. Gasser C, Taiber S, Yeh CM, Wittig CH, Hegemann P, Ryu S, Wunder F, Möglich A: Engineering of a red-light-activated human cAMP/cGMP-specific phosphodiesterase. Proc Natl Acad Sci U S A 2014, 111:8803-8808. The activity of a fusion between the photosensory module of DrBphP and a cAMP/cGMP phosphodiesterase can be light-regulated by a factor of six. This suffices already to control many cAMP/cGMP-dependent signaling pathways in eukaroytes. 49. Singer P, Fey S, Goller AH, Hermann G, Diller R: Femtosecond dynamics in the lactim tautomer of phycocyanobilin: a longwavelength absorbing model compound for the phytochrome chromophore. ChemPhysChem 2014, 15:3824-3831. 50. Falklöf O, Durbeej B: Modeling of phytochrome absorption spectra. J Comput Chem 2013, 34:1363-1374. Current Opinion in Structural Biology 2015, 35:7–16 56. Song C, Psakis G, Lang C, Mailliet J, Gärtner W, Hughes J, Matysik J: Two ground state isoforms and a chromophore D-ring photoflip triggering extensive intramolecular changes in a canonical phytochrome. Proc Natl Acad Sci U S A 2011, 108:3842-3847. 57. Kim PW, Rockwell NC, Martin SS, Lagarias JC, Larsen DS: Heterogeneous photodynamics of the Pfr state in the cyanobacterial phytochrome Cph1. Biochemistry 2014, 53:4601-4611. 58. Diensthuber RP, Bommer M, Gleichmann T, Möglich A: Fulllength structure of a sensor histidine kinase pinpoints coaxial coiled coils as signal transducers and modulators. Structure 2013, 21:1127-1136. 59. Müller K, Engesser R, Timmer J, Nagy F, Zurbriggen MD, Weber W: Synthesis of phycocyanobilin in mammalian cells. Chem Commun 2013, 49:8970-8972. Overexpression of heme oxygenase 1 and PcyA from T. elongatus in the mitochondria of CHO cells allows to achieve cytosolic PCB levels of up to 2 mM, which allow to form holo-phytochromes in mammals without exogenously added bilin chromophore. 60. Zhang J, Wu X-J, Wang Z-B, Chen Y, Wang X, Zhou M, Scheer H, Zhao K-H: Fused-gene approach to photoswitchable and fluorescent biliproteins. Angew Chem Int Ed Engl 2010, 49:5456-5458. www.sciencedirect.com