ubiquitinatin
advertisement

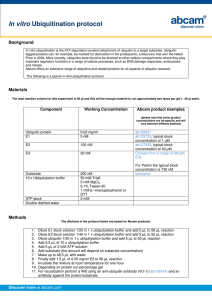
Post-translational Modification Ubiquitination Introduction • Goldknopf and his friends realized that histones can be modified by the protein ubiquitin through Lys-linked isopeptide bonds. • The prevalence and importance of protein- based modifications emerged in the 1980s, when landmark studies connected the ATP-dependent ubiquitination of substrates to their degradation by the 26S proteasome • protein function can be regulated through chemical modifications of amino acids Ubiquitin….? First identified in 1975 A 8.6 kDa protein Expressed in all eukaryotic cells and highly conserved A 76-amino acid protein Functions of ubiquitin and ubiquitination pathway were elucidated in the early 1980s by Aaron Ciechanover, Avram Hershko, and Irwin Rose o Awarded Nobel Prize in Chemistry in 2004 In mammals encoded by 4 different genes UBA52 and RPS27A genes ---monoubiquitin UBB and UBC genes--polyubiquitin precursor proteins Ubiquitin….? Key features-seven Lys residues and C-terminal tail Ubiquitin….? Recognized by a hydrophobic surface that consists of Ile44, Leu8, Val70, and His68 figure • The Ile44 ---bound by the proteasome and most UBDs ---essential for cell division Another hydrophobic surface is centered on Ile36 and involves Leu71 and Leu73 figure • The Ile36 ---mediate ubiquitin-ubiquitin interactions ---recognized by HECTE3s, DUBs and UBDs Recognized through C-terminal tail figure • β1/β2 loop containing Leu8 shows flexibility o important for recognition by ubiquitin-binding proteins Ubiquitin….? Insert 3 figures for each structure Ubiquitination • Ubiquitination (or ubiquitylation) is an enzymatic posttranslational modification in which an ubiquitin protein is attached to a substrate protein • This process most commonly binds the last amino acid of ubiquitin (glycine 76) to a lysine residue on the substrate. • An isopeptide bond is formed between the carboxyl group (COO−) of the ubiquitin's glycine and the epsilon-amino group (ε-NH+3) of the substrate's lysine • Trypsin cleavage of a ubiquitin-conjugated substrate leaves a di-glycine "remnant" that is used to identify the site of ubiquitination. Ubiquitination • Adding ubiquitin to electron-rich nucleophiles, sites in a protein -----"non-canonical ubiquitination". o The amine group N-terminus of the protein MyoD, 22 other proteins in multiple species and also in ubiquitin itself o the sulfhydryl group on cysteine, and o the hydroxyl group on threonine and serine. • Ubiquitination requires three types of enzyme: o E1s ubiquitin-activating enzymes o E2s ubiquitin-conjugating enzymes o E3s ubiquitin ligases– many(hundreds) Ubiquitination • Ubiquitin is attached to substrates by a complex 3-step cascades 1) Activation E1 (ubiquitin-activating enzyme) ATP dependent two-step reaction 1st --- production of a ubiquitin-adenylate intermediate. The E1 binds both ATP and ubiquitin and catalyses the acyl-adenylation of the C-terminus of the ubiquitin molecule. 2nd transfers ubiquitin to an active site cysteine residue, with release of AMP. This step results in a thioester linkage between the C-terminal carboxyl group of ubiquitin and the E1 cysteine sulfhydryl group. The human genome contains two genes: UBA1 and UBA6 Ubiquitination 2) Conjugation E2 (ubiquitin-conjugating enzymes) catalyze the transfer of ubiquitin from E1 to the active site cysteine of the E2 via a trans(thio)esterification reaction. In order to perform this reaction, the E2 binds to both activated ubiquitin and the E1 enzyme. Humans possess 35 different E2 enzymes, whereas other eukaryotic organisms have between 16 and 35 E2 have UBC (ubiquitin-conjugating catalytic) fold which is highly conserved structure Ubiquitination 3) Ligation E3 (ubiquitin ligases) Create an isopeptide bond between a lysine of the target protein and the C-terminal glycine of ubiquitin. E3 enzymes function as the substrate recognition modules of the system can Interact with both E2 and substrate some activate the E2 enzymes E3 enzymes possess one of two domains: Homologous to the E6-AP carboxyl terminus (HECT) domain --transiently bind ubiquitin (thioester intermediate is formed with the active-site cysteine of the E3) Really interesting new gene (RING) domain (or U-box domain) --catalyze the direct transfer from the E2 enzyme to the substrate Ubiquitination • In the ubiquitination cascade, E1 can bind with many E2s, which bind with hundreds of E3s in a hierarchical way. • E4 enzymes (ubiquitin-chain elongation factors) o adding pre-formed polyubiquitin chains to substrate proteins o For example, addition of a polyubiquition chain using p300 and CBP on monoubiquitylated tumor suppressor p53 • The end result of this process is o the addition of one ubiquitin molecule (monoubiquitination) or o a chain of ubiquitin molecules (polyubiquitination) to the substrate protein. Monoubiquitination • Multi-monoubiquitination is the addition of one ubiquitin molecule to multiple substrate residues. • is thought to be required prior to the formation of polyubiquitin chains. • Affects cellular processes such as o Endocytosis o Membrane trafficking Polyubiquitination • Formation of a ubiquitin chain on a single lysine residue on the substrate protein • These chains are made by linking the glycine residue of a ubiquitin molecule to a lysine of ubiquitin bound to a substrate. • Ubiquitin is attached to the N-terminus of a 2nd ubiquitin fig • Ubiquitin has 7-lysine residues and an N-terminus that serves as points of ubiquitination; they are • Lys48-linked chains---predominant (> 50% of all linkages) o Role --- target proteins to the 26s proteasome for degradation(at least four) Polyubiquitination • Lys63- linked chains---second abundant via performs various non-degradative roles by binding with ESCRT-0 o Endocytic trafficking, inflammation, translation, and DNA repair • Remaining Lys6, Lys11, Lys27, Lys29 or Lys33 and Met1 o chains linked to this parts can induce proteasomal degradation • Researches to characterize this remaining residues led to the discovery of highly linkage-specific enzymes and proteins that assemble, recognize and hydrolyze each ubiquitin chain type, i.e., ‘write’, ‘read’ and ‘erase’ this ubiquitin code Polyubiquitination • Structure of linked chains have specific effects on the protein • Lys29-, Lys33-, Lys63- & M1-linked chains form open(linear) conformation chains fig o do not interact with each other, except for the covalent isopeptide bonds linking them together • Lys6-, Lys11-, and Lys48-linked chains form closed conformations fig o have interfaces with interacting residues • Proteins can specifically bind to ubiquitin via ubiquitin-binding domains (UBDs) Polyubiquitination • The distances between individual ubiquitin units in chains differ between lysine 63- and 48-linked chains. • The UBDs exploit this by having o small spacers between ubiquitin-interacting motifs that bind lysine 48-linked chains o larger spacers between ubiquitin-interacting motifs that bind lysine 63-linked chains • The machinery involved in recognizing polyubiquitin chains can also differentiate between Lys63-linked chains and M1- (induce proteasomal degradation) linked chains, Function of Ubiquitination Transcriptional regulation Histones---usually (mono-), rarely (poly)-ubiquitinated o alters chromatin structure and allows the access of enzymes involved in transcription. o acts as a binding site for proteins that either activate or inhibit transcription o These effects can modulate the transcription of genes Protein regulation Comprehensive proteomics studies identified tens-ofthousands of ubiquitination sites on thousands of proteins. Suggests most proteins will experience ubiquitination at some point in their cellular lifetime Function of Ubiquitination Membrane proteins Multi-monoubiquitination o Mark transmembrane proteins for removal from membranes (eg. Receptors)-----How? • When tagged with ubiquitin, • the protein subcellular localization is altered • targeting for destruction in lysosomes • a negative feedback mechanism because often the stimulation of receptors by ligands increases their rate of ubiquitination and internalisation lysine 63-linked polyubiquitin chains o in the trafficking some membrane proteins Function of Ubiquitination Genomic maintenance Proliferating cell nuclear antigen (PCNA) ---protein involved in DNA synthesis Under normal physiological conditions PCNA is sumoylated When DNA is damaged by ultra-violet radiation or chemicals, the SUMO molecule that is attached to a lysine residue is replaced by ubiquitin Monoubiquitinated PCNA recruits polymerases for repair but prone to error resulting in the synthesis of mutated DNA Lysine 63-linked polyubiquitination of PCNA allows a less error-prone • bypass mutation by the template switching pathway. Function of Ubiquitination Infection and immunity • Regulate immune signal transduction pathways at all stages o o o o o steady-state repression, activation during infection and attenuation upon clearance Without this regulation resulting in chronic disease or death Alternatively hyper-activate the immune system resulting autoimmune damage in organs and tissues • Viruses can easily overcome host cells by blocking the ubiquitin, system which have high role in cells, to support their own replication • In humans the retinoic acid-inducible gene I (RIG-I) protein o primary immune system sensor for viral and other invasive RNA Malfunction of Ubiquitination Genetic disorders • Neurodegenerative disorders: Alzheimer's and Parkinson's o lesions of different transcripts of ubiquilin-1. o Higher levels of ubiquilin in the braindecrease malformation of amyloid precursor protein (APP) trigger Alzheimer's disease o lower levels of ubiquilin-1 in the brain have been associated with increased malformation of APP • A frameshift mutation in ubiquitin B abnormal peptide, UBB+1 accumulation in Alzheimer's disease • Angelman syndrome is caused by a disruption of UBE3A, which encodes a ubiquitin E3 ligase enzyme • 3-M syndrome is an autosomal-recessive growth retardation disorder associated with mutations of the Cullin7 E3 ubiquitin ligase Increased ubiquitination activity Cervical cancer Human papillomavirus (HPV) are known to hijack cellular ubiquitin-proteasome pathway for viral infection and replication. 1. The E6 proteins of HPV will bind to the N-terminus of the cellular E6-AP E3 ubiquitin ligase, 2. Redirecting the complex to bind p53, a well-known tumor suppressor gene that inactivation is found in many types of cancer 3. p53 undergoes ubiquitination and proteasome-mediated degradation. 4. The E7 proteins of HPV will bind to Rb, also a tumor suppressor gene, mediating its degradation. Loss of p53 and Rb in cells allows limitless cell proliferation Increased ubiquitination activity p53 regulation • MDM2 a gene encodes for a RING E3 Ubiquitin ligase • downregulation of p53 activity • MDM2 targets p53 for ubiquitination and proteasomal degradation thus keeping normal cell condition • Overexpression of MDM2 causes loss of p53 activity and therefore allowing cells to have a limitless replicative potential Efp (estrogen-inducible RING-finger protein) • E3 ubiquitin ligase • Its overexpression has been shown to be the major cause of estrogen-independent breast cancer Deubiquitination • Deubiquitinating enzymes (DUBs) • Removing ubiquitin from substrate protein • They are cysteine proteases that cleave the amide bond between the two proteins • They are highly specific • They can cleave both isopeptide (between ubiquitin and lysine) and peptide bonds (between ubiquitin and the N-terminus). • Cleave polyubiquitin to produce active ubiquitin or form monoubiquitin • Recycle ubiquitin that has been bound to small nucleophilic molecules during the ubiquitination process. Ubiquitination Further ubiquitin modifications • Ubiquitin is also be modified by other modifications. E.g. o by SUMO (small ubiquitin-like modifier family) • SUMOylation of (poly)ubiquitin is conceptually similar to polyubiquitin chain formation • Ubiquitin is also modified by acetylation • Mining of available datasets indicates that 6 out of 7 ubiquitin Lys residues can become acetylated Further ubiquitin modifications • Ubiquitin phosphorylation on---• provide additional regulation in the ubiquitin system • For these modifications o Writers--- kinases and acetyltransferases, o Eraser--- phosphatases and deacetylases and o Readers--- phospho-ubiquitin (phosphoUb) or acetylubiquitin (AcUb) binding domains Human and Yeast Ub ----highly conserved