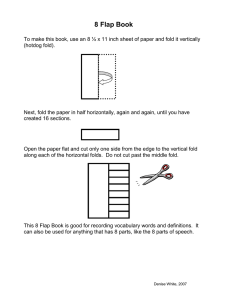
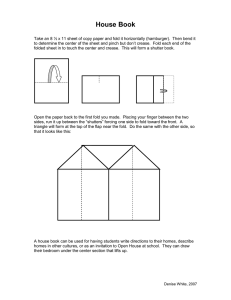
Toll-Like.ppt
advertisement
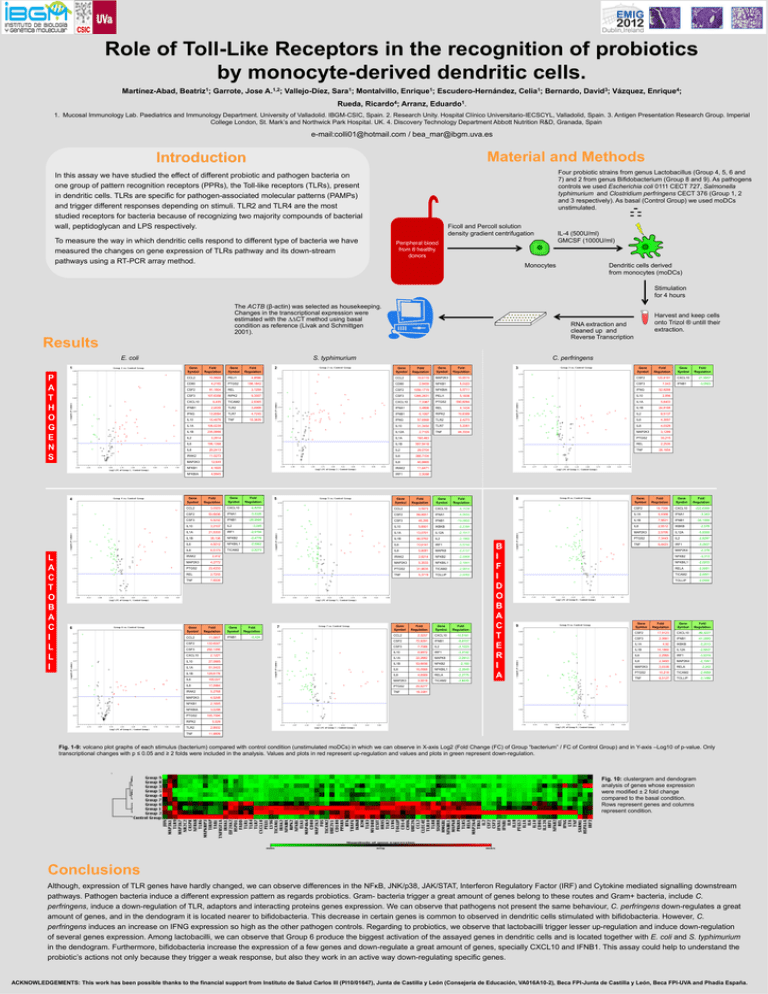
Role of Toll-Like Receptors in the recognition of probiotics by monocyte-derived dendritic cells. Martínez-Abad, Beatriz1; Garrote, Jose A.1,2; Vallejo-Díez, Sara1; Montalvillo, Enrique1; Escudero-Hernández, Celia1; Bernardo, David3; Vázquez, Enrique4; Rueda, Ricardo4; Arranz, Eduardo1. 1. Mucosal Immunology Lab. Paediatrics and Immunology Department. University of Valladolid. IBGM-CSIC, Spain. 2. Research Unity. Hospital Clínico Universitario-IECSCYL, Valladolid, Spain. 3. Antigen Presentation Research Group. Imperial College London, St. Mark’s and Northwick Park Hospital. UK. 4. Discovery Technology Department Abbott Nutrition R&D, Granada, Spain e-mail:colli01@hotmail.com / bea_mar@ibgm.uva.es Material and Methods Introduction Four probiotic strains from genus Lactobacillus (Group 4, 5, 6 and 7) and 2 from genus Bifidobacterium (Group 8 and 9). As pathogens controls we used Escherichia coli 0111 CECT 727, Salmonella typhimurium and Clostridium perfringens CECT 376 (Group 1, 2 and 3 respectively). As basal (Control Group) we used moDCs unstimulated. In this assay we have studied the effect of different probiotic and pathogen bacteria on one group of pattern recognition receptors (PPRs), the Toll-like receptors (TLRs), present in dendritic cells. TLRs are specific for pathogen-associated molecular patterns (PAMPs) and trigger different responses depending on stimuli. TLR2 and TLR4 are the most studied receptors for bacteria because of recognizing two majority compounds of bacterial wall, peptidoglycan and LPS respectively. To measure the way in which dendritic cells respond to different type of bacteria we have measured the changes on gene expression of TLRs pathway and its down-stream pathways using a RT-PCR array method. Ficoll and Percoll solution density gradient centrifugation IL-4 (500U/ml) GMCSF (1000U/ml) Peripheral blood from 6 healthy donors Monocytes Dendritic cells derived from monocytes (moDCs) Stimulation for 4 hours The ACTB (β-actin) was selected as housekeeping. Changes in the transcriptional expression were estimated with the ∆∆CT method using basal condition as reference (Livak and Schmittgen 2001). RNA extraction and cleaned up and Reverse Transcription Results E. coli 1 P A T H O G E N S Gene Symbol 10,9988 PELI1 CD80 4,2165 PTGS2 CSF2 91,1504 CSF3 107,6358 6,435 Fold Regulation 2 C. perfringens Gene Symbol Fold Regulation Gene Symbol Fold Regulation MAP2K3 10,8515 NFKB1 3 Gene Symbol Fold Regulation Gene Symbol Fold Regulation CSF2 120,8161 CXCL10 -21,8607 5,0323 CSF3 7,543 52,9298 4,8596 CCL2 10,6119 198,1842 CD80 2,9459 REL 3,1259 CSF2 1056,1719 NFKBIA 5,5711 IFNG RIPK2 9,3557 CSF3 1266,2631 PELI1 5,1836 IL10 2,856 TICAM2 2,8365 CXCL10 7,3387 PTGS2 590,6094 IL1A 5,6403 4,1434 IL1B 24,8188 IFNB1 2,4435 TLR2 3,4999 IFNA1 3,4806 REL IFNG 13,8084 TLR7 4,7245 IFNB1 6,1087 RIPK2 10,8368 IL2 8,5137 IL10 10,4676 TNF 15,3839 IFNG 57,6998 TLR2 2,4273 IL6 4,3657 IL1A 106,0239 IL10 31,3454 TLR7 5,2061 IL8 4,0329 IL1B 235,5998 IL12A 2,7105 TNF 44,3334 MAP2K3 3,1299 PTGS2 33,215 IL2 3,2814 IL1A 193,463 IL6 196,1398 IL1B 587,9418 REL 2,2535 IL8 20,2413 IL2 29,0704 TNF 28,1654 IRAK2 11,5273 IL6 388,7104 MAP2K3 12,649 IL8 40,6665 NFKB1 4,1605 IRAK2 11,6471 NFKBIA 4,6945 IRF1 Gene Symbol Fold Regulation Gene Symbol Fold Regulation CCL2 3,0323 CXCL10 -6,8258 CSF2 50,6936 IFNA1 CSF3 4,5232 IFNB1 IL10 3,2107 IL2 IL1A 21,5333 IL1B 5 Fold Regulation CSF2 18,7006 CXCL10 -222,6368 -4,0655 IL1A 4,4368 IFNA1 -3,343 IFNB1 -19,0858 IL1B 7,9621 IFNB1 -34,1568 5,6801 IKBKB -2,2394 IL8 2,5512 IKBKB -2,578 IL1A 13,0701 IL12A -2,1917 MAP2K3 2,5706 IL12A -5,8398 -2,4778 IL1B 46,0762 IL2 -2,1888 PTGS2 7,3443 IL2 -2,8297 NFKBIL1 -2,5962 IL6 13,6141 IRF1 -3,5744 TNF 6,4423 IRF1 -3,2827 TICAM2 -2,5273 IL8 5,6081 MAPK8 -2,4137 IRAK2 2,8214 NFKB2 -2,0968 MAP2K3 5,3533 NFKBIL1 -2,1841 31,9635 TICAM2 -2,9914 5,3719 TOLLIP -2,0092 CCL2 3,5973 CXCL10 -4,1539 -3,5326 CSF2 68,8851 IFNA1 -28,9985 CSF3 46,295 -3,045 IL10 IRF1 -2,9194 35,136 NFKB2 IL6 4,5012 IL8 6,5173 4,2772 23,4233 2,7255 TNF 7,8538 Gene Symbol CCL2 CSF2 CSF3 CXCL10 Gene Symbol 2,412 REL Fold Regulation 11,5807 132,6047 PTGS2 TNF Gene Symbol IFNB1 Fold Regulation -5,424 7 Gene Symbol Fold Regulation Gene Symbol Fold Regulation CCL2 2,3257 CXCL10 -10,5161 CSF2 72,8291 CSF3 IFNB1 -9,8707 7,7066 IL2 -2,1033 IL10 6,9872 IRF1 -3,3142 IL1A 22,2682 MAPK8 -2,0913 IL1B 53,6656 NFKB2 -2,165 IL6 16,0568 NFKBIL1 -2,3649 IL8 4,6569 RELA -2,2775 3,5618 TICAM2 -2,8438 292,1396 2,1221 IL10 27,0685 IL1A 51,0403 IL1B 129,6176 IL6 100,027 MAP2K3 IL8 17,5964 PTGS2 25,5277 TNF 18,3381 IRAK2 5,2768 MAP2K3 4,5248 NFKB1 2,1695 NFKBIA 3,0298 PTGS2 8 Gene Symbol Fold Regulation PTGS2 -3,0923 Fold Regulation Gene Symbol MAP2K3 IFNB1 2,3088 Fold Regulation IRAK2 6 Fold Regulation CCL2 CXCL10 4 L A C T O B A C I L L I S. typhimurium Gene Symbol Harvest and keep cells onto Trizol ® untill their extraction. B I F I D O B A C T E R I A 9 Gene Symbol Gene Symbol MAP2K4 -2,378 NFKB2 -3,313 NFKBIL1 -2,0203 RELA -2,0651 TICAM2 -2,0991 TOLLIP -2,0556 Fold Regulation Gene Symbol Fold Regulation CSF2 17,9123 CXCL10 -89,3227 CSF3 2,3981 IFNB1 -41,2893 IL1A 4,92 IKBKB -2,2312 IL1B 14,1869 IL12A -2,9637 IL6 2,2965 IRF1 -3,5319 IL8 2,3493 MAP2K4 -2,1041 MAP2K3 2,0338 RELA PTGS2 10,218 TICAM2 -2,4988 TNF 6,5127 TOLLIP -2,1489 -2,202 105,1594 RIPK2 5,529 TLR2 2,8932 TNF 11,9809 Fig. 1-9: volcano plot graphs of each stimulus (bacterium) compared with control condition (unstimulated moDCs) in which we can observe in X-axis Log2 (Fold Change (FC) of Group “bacterium” / FC of Control Group) and in Y-axis –Log10 of p-value. Only transcriptional changes with p ≤ 0.05 and ≥ 2 folds were included in the analysis. Values and plots in red represent up-regulation and values and plots in green represent down-regulation. Fig. 10: clustergram and dendogram analysis of genes whose expression were modified ± 2 fold change compared to the basal condition. Rows represent genes and columns represent condition. Conclusions Although, expression of TLR genes have hardly changed, we can observe differences in the NFκB, JNK/p38, JAK/STAT, Interferon Regulatory Factor (IRF) and Cytokine mediated signalling downstream pathways. Pathogen bacteria induce a different expression pattern as regards probiotics. Gram- bacteria trigger a great amount of genes belong to these routes and Gram+ bacteria, include C. perfringens, induce a down-regulation of TLR, adaptors and interacting proteins genes expression. We can observe that pathogens not present the same behaviour, C. perfringens down-regulates a great amount of genes, and in the dendogram it is located nearer to bifidobacteria. This decrease in certain genes is common to observed in dendritic cells stimulated with bifidobacteria. However, C. perfringens induces an increase on IFNG expression so high as the other pathogen controls. Regarding to probiotics, we observe that lactobacilli trigger lesser up-regulation and induce down-regulation of several genes expression. Among lactobacilli, we can observe that Group 6 produce the biggest activation of the assayed genes in dendritic cells and is located together with E. coli and S. typhimurium in the dendogram. Furthermore, bifidobacteria increase the expression of a few genes and down-regulate a great amount of genes, specially CXCL10 and IFNB1. This assay could help to understand the probiotic’s actions not only because they trigger a weak response, but also they work in an active way down-regulating specific genes. ACKNOWLEDGEMENTS: This work has been possible thanks to the financial support from Instituto de Salud Carlos III (PI10/01647), Junta de Castilla y León (Consejería de Educación, VA016A10-2), Beca FPI-Junta de Castilla y León, Beca FPI-UVA and Phadia España.