samsi
advertisement

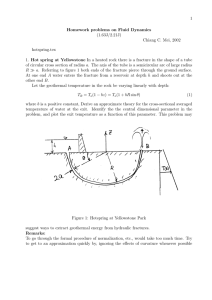
Random Fields of Multivariate Test Statistics, with Applications to “Shape” Analysis Keith Worsley, McGill Jonathan Taylor, Stanford and Université de Montréal Arnaud Charil, Montreal Neurological Institute Francesco Tomaiuolo Fondazione ‘Santa Lucia’, Roma Are Multiple Sclerosis lesions related to patient disability? Data: n = 425 mild MS patients Disability measured by EDSS and other scores 8 7 6 Correlation = 0.290, T = 6.23 (423 df) 5 EDSS 4 3 2 1 0 0 10 20 30 40 50 60 Total lesion volume (cc) 70 80 Charil et al, NeuroImage (2003) Which Multiple Sclerosis lesions are related to patient disability? Repeat for every voxel: standard VBM study Find lesion density at every voxel: Correlation(Lesion density, EDSS) Segment MS lesions: 1=lesion, 0=not Smooth 10mm Convert to T statistic (423 df) Threshold? Bonferroni? Too conservative Random field theory …  ¹= 0·µ·¼=2 Example test statistic: Chi-bar max Z1 cos µ + Z2 sin µ Z1~N(0,1) Z2~N(0,1) s2 3 2 1 0 -1 -2 Excursion sets, Xt = fs :  ¹ ¸ tg s1 -3 Threshold t 4 Rejection regions, Z2 2 Search Region, S Rt = fZ :  ¹ ¸ tg 3 2 1 0 Z1 -2 -2 0 2 Euler characteristic heuristic: EC = #blobs - # holes Search Region, S Euler characteristic, EC EC= 1 7 Excursion sets, Xt 6 5 2 1 1 0 10 Observed 8 ¸ t) P(max Â(s) ¹ s2S 6 ¼ E(EC) = 0:05 Expected ) t = 3:75 4 2 0 -2 0 0.5 EXACT! 1 1.5 2 E(EC(S \ Xt )) = X D d=0 2.5 3 L (S)½ (t) d d 3.5 4 Threshold, t E(EC(S \ X )) = t Beautiful symmetry: X D L (S)½ (t) d d d=0 Steiner-Weyl Tube Formula (1930) Tube(λS,r) Taylor Gaussian Tube Formula (2003) Radius, r Tube(Rt,r) Z2 14 µ ¶ @Z ¸ = Sd p @s 4 log 2 = FWHM r 12 2 10 2 r 1.5 1 Rt 0.8 0.6 λS 8 Radius, r 0 1 6 4 0.5 2 Z1 0.4 0.2 -2 L (S)0 EC density ½ (t) Lipschitz-Killing2 curvature 4 6 8 10 12 14 d -2 0 2d jTube(¸S; r)j L (S) 2 L 2 1 (S)r ¼L0 (S)r2 Area 100 50 0 jTube(¸S; r)j = X D d=0 0 0.5 1 1.5 ¡(d=2 + 1) L D ¡d 0.3 (S)r d p 2¼½1 (t)r ½0 (t) ¼½2 (t)r2 0.2 0.1 0 2 Radius of Tube, r ¼d P(Tube(Rt ; r)) 0.4 Probability 150 0 0 0.5 Radius of Tube, r Adler & Taylor, Rarndom Fields and Geometry (2007) 1 P(Tube(Rt ; r)) = 1 X (2¼)d=2 d! d=0 ½d (t)rd EC density ½d (t) of the  ¹ statistic Z2~N(0,1) r Tube(Rt,r) Rejection region Rt t-r t Taylor’s Gaussian Tube Formula: 1 X P (Z1 ; Z2 2 Tube(Rt ; r)) = Z1~N(0,1) (2¼)d=2 ½d (t)rd d! d=0 ½0 (t) = Z = ½0 (t) + (2¼)1=2 ½1 (t)r + (2¼)½2 (t)r2 =2 + ¢ ¢ ¢ Z 1 = (2¼)¡1=2 e¡z2 =2 dz + e¡(t¡r)2 =2 =4 t¡r 1 (2¼)¡1=2 e¡z2 =2 dz + e¡t2 =2 =4 t ½1 (t) = (2¼)¡1 e¡t2 =2 + (2¼)¡1=2 e¡t2 =2 t=4 ½2 (t) = (2¼)¡3=2 e¡t2 =2 t + (2¼)¡1 e¡t2 =2 (t2 ¡ 1)=8 .. . Taylor & Worsley, Annals of Statistics, submitted (2007) Lipschitz-Killing curvature Ld (S) r Tube(λS,r) λS µ ¶ @Z ¸ = Sd p @s 4 log 2 = FWHM Steiner-Weyl Volume of Tubes Formula: Area(Tube(¸S; r)) = X D ¼ d=2 L ¡d (S)r d D ¡(d=2 + 1) d=0 = L2 (S) + 2L1 (S)r + ¼ L0 (S)r2 = Area(¸S) + Perimeter(¸S)r + EC(¸S)¼r2 L (S) = EC(¸S) = Resels0 (S) 0 p L (S) = 1 Perimeter(¸S) = 4 log 2 Resels (S) 1 1 2 L (S) = Area(¸S) = 4 log 2 Resels2 (S) 2 Lipschitz-Killing curvatures are just “intrinisic volumes” or “Minkowski functionals” in the metric of the variance of the derivative of the process How to ¯nd Lipschitz-Killing curvature Ld (S) S S S ¸ = Sd Edge length × λ FWHM/√(4log2) 12 10 8 6 4 2 . .. . . . . . . .. . . .. . . . . . . . . . . . . . . . .. . . . . . ... . . 4 .. . . . . . . . . . . . 6 .. . . . . . . . . . . . . . . . . . . . . . . . 8 .. . . . ... . .. . . . . . . . ..... . . . . .... .. .. 10 µ @Z @s ¶ p = 4 log 2 FWHM of simplices L (Lipschitz-Killing ²) = 1, L (¡) curvature L (N = 1, )=1 0 0 0 L (¡) = edge length, L (N) = 1 perimeter 1 2 L1 (N) = area 2 P Lcurvature P L Lipschitz-Killing union L ² ¡ Pof L ¡ of simplices N (S) = P² 0 ( ) ¡ 0( ) + P L (S) = L (¡) ¡ L (N) ¡ N 1 L1 (S) = P L 1(N) 2 N 2 0 N 0 ( ) p Var(∇Z) Non-isotropic data? Use Riemannian metric µ ¶ of ¸ = Sd Z~N(0,1) s2 3 @Z @s = 4 log 2 FWHM 2 1 0.14 0.12 0 -1 -2 Edge length × λ FWHM/√(4log2) 12 10 8 6 4 2 .. . . . . . . . . .. . . .. . . . . . . . . . . . . . . . .. . . . . . ... . . . . . . . . . . . . . . 4 6 .. . . . . . . . . . . . . . . . . . . . . . . . 8 . .. . . . ... . .. . . . . . . . ..... . . . . .... ... 10 0.1 0.08 0.06 -3 s1 of simplices L (Lipschitz-Killing ²) = 1, L (¡) curvature L (N = 1, )=1 0 0 0 L (¡) = edge length, L (N) = 1 perimeter 1 2 L1 (N) = area 2 P Lcurvature P L Lipschitz-Killing union L ² ¡ Pof L ¡ of simplices N (S) = P² 0 ( ) ¡ 0( ) + P L (S) = L (¡) ¡ L (N) ¡ N 1 L1 (S) = P L 1(N) 2 N 2 0 N 0 ( ) Estimating Lipschitz-Killing curvature Ld (S) We need independent & identically distributed random fields e.g. residuals from a linear model Z1 Z2 Z3 Z4 Z5 Z6 Z7 Z9 … Zn Z8 Replace coordinates of the simplices in S⊂RealD by normalised residuals (Z1,…,Zn) / ||(Z1,…,Zn)|| in Realn of simplices L (Lipschitz-Killing ²) = 1, L (¡) curvature L (N = 1, )=1 0 0 0 L (¡) = edge length, L (N) = 1 perimeter 1 2 L1 (N) = area 2 P Lcurvature P L Lipschitz-Killing union L ² ¡ Pof L ¡ of simplices N (S) = P² 0 ( ) ¡ 0( ) + P L (S) = L (¡) ¡ L (N) ¡ N 1 L1 (S) = P L 1(N) 2 N 2 0 N 0 Unbiased! ( ) Unbiased! Taylor & Worsley, JASA (2007) Which Multiple Sclerosis lesions are related to patient disability? L (S) = 0 0 L (S) = 79:1 1 L (S) = 588:6 2 L (S) = 1404:1 3 º = 423 Z 1 ½0 (t) = ¢ µ ¶¡(º+1)=2 2 ¡ º+1 u 2 ¡ ¢ 1+ du º º 1=2 (º¼) ¡ t 2 µ ¶ ¡(º ¡1)=2 2 t ½1 (t) = (2¼)¡1 1 + º ¡ ¢ µ ¶¡(º ¡1)=2 º+1 ¡ t2 ¡3=2 2 ½2 (t) = (2¼) ¡ ¢ ¡ ¢ t 1+ º 1=2 º ¡ º 2 µ 2¡ ¶µ ¶¡(º ¡1)=2 2 º 1 ¡ t ½3 (t) = (2¼)¡2 t2 1 1+ º º P(max T (s) ¸ t) ¼ E(EC(S \ Xt )) = s2S ¡ X 3 d=0 L (S)½ (t) = 0:05; t = 4:47 d d MS lesions and cortical thickness Idea: MS lesions interrupt neuronal signals, causing thinning in down-stream cortex Average cortical thickness (mm) 5.5 5 4.5 4 3.5 3 2.5 Correlation = -0.568, T = -14.20 (423 df) 2 1.5 0 10 20 30 40 50 Total lesion volume (cc) 60 70 80 Charil et al, NeuroImage (2007) MS lesions and cortical thickness at all pairs of points Dominated by total lesions and average cortical thickness, so remove these effects Cortical thickness, smoothed 20mm Lesion density, smoothed 10mm Find partial correlation(lesion density, cortical thickness) removing total lesion volume linear model: CT-av(CT) ~ 1 + TLV + LD, test for LD Repeat of all voxels in 3D, nodes in 2D Subtract average cortical thickness ~1 billion correlations, so thresholding essential! Look for high negative correlations … Thresholding? Correlation random field Correlation between 2 fields at 2 different locations, searched over all pairs of locations one in R (D dimensions), one in S (E dimensions) µ ¶ X D X E ¼ L (R) L (S) ½ (c) P max Correlation > c d e d;e R;S d=0 e=0 X (d ¡ 1)!e!2n¡d¡e¡2 X ¡ ¡d¡e¡1 ¡ ¡ ¡ n ½d;e (c) = ( 1)k cd+e 1 2k (1 c2 ) 2 +k +1 ¼ d+e 2 k i;j ¡( n¡d + i)¡( n¡e + j) 2 2 ¡ ¡ ¡ ¡ ¡ i!j!(k i j)!(n 1 d e + i + j + k)!(d ¡ 1 ¡ k ¡ i + j)!(e ¡ k ¡ j + i)! Cao & Worsley, Annals of Applied Probability (1999) MS lesion data: P=0.05, c=0.300, T=6.46 Cluster extent rather than peak height (Friston, 1994) fit a quadratic Y to the peak: L (cluster) »c ® D Peak height Choose a lower level, e.g. t=3.11 (P=0.001) Find clusters i.e. connected components of excursion set Z D=1 Measure cluster L extent extent by D t Distribution: k Distribution of maximum cluster extent: s Bonferroni on N = #clusters ~ E(EC). Cao, Advances in Applied Probability (1999) How do you choose the threshold t for defining clusters? If signal is focal i.e. ~FWHM of noise If signal is broad i.e. >>FWHM of noise Choose a high threshold i.e. peak height is better Choose a low threshold i.e. cluster extent is better Conclusion: cluster extent is better for detecting broad signals Alternative: smooth data with filter that matches signal (Matched Filter Theorem)… try range of filter widths … scale space search … correct using random field theory … a lot of work … Cluster extent is easier! Multivariate linear models for random field data Y(s) = X B(s) + E(s), test H0: B(s)=0, s in S nxq nxp pxq nxq EC densities known (<2007) for: q=1 p=1 p>1 T F q>1 Hotelling’s T2 Which to choose? Wilks’ Λ ? Pillai’s trace ? Roy’s max root ? but not these! Roy’s union-intersection principle (1954) Make it into a univariate linear model by multiplying by vector qx1 v Y(s)v = X B(s)v + E(s)v, H0: B(s)v=0 nx1 P µ nxp px1 nx1 Calculate usual F-statistic F(s,v) Maxv in unit q-sphere V F(s,v) = Roy’s maximum root Now it is an F-field in search region S ⊗ V and we already know¶ the EC density of the F-field: max F (s; v) ¸ t s2S;v 2V ¼ D+q X d=0 L (S d V )½F (t) = d X D d=0 L (S) d q X L (V )½F k d+k (t) k=0 Taylor & Worsley, Annals of Statistics (2007) Maximum canonical correlation Let X(s), s 2 S ½ <M , and Y (t), t 2 T ½ <N be matrices of Gaussian random ¯elds with p and q columns and the same number º of rows. De¯ne the maximum canonical correlation random ¯eld as u0 X(s)0 Y (t)v C(s; t) = max u;v (u0 X(s)0 X(s)u v0Y (t)0 Y (t)v)1=2 ; the maximum of the canonical correlations between X and Y , de¯ned as the singular values of (X 0 X)¡1=2 X 0 Y (Y 0 Y )¡1=2 . P µ max C(s; t) ¸ c s2S;t2T ¶ ¼ 1 2 X M i=0 L (S) i X N L (T ) j j=0 p X L (U ) k k=0 q X L (V )½ (c) l i+k;j+l l=0 where U is the unit sphere in <p , V is the unit sphere in <q . ¡ ¢ k+1 ¼ k ¡ p+1 2 L (U ) = ³ 2 ´2 k ¡ ¡ k! p 1 k ! 2 if p ¡ 1 ¡ k is even, and zero otherwise, k = 0; : : : ; p ¡ 1. Now available in stat_threshold.m Deformation Based Morphometry (DBM) (Tomaiuolo et al., 2004) n1 = 19 non-missile brain trauma patients, 3-14 days in coma, n2 = 17 age and gender matched controls Data: non-linear vector (q=3) deformations needed to warp each MRI to an atlas standard Locate damage: find regions where deformations are different, hence shape change Is damage connected? Find pairs of regions with high canonical correlation. Worsley et al. NeuroImage (2004)