RBD_Subsampling_sep192008.doc
advertisement
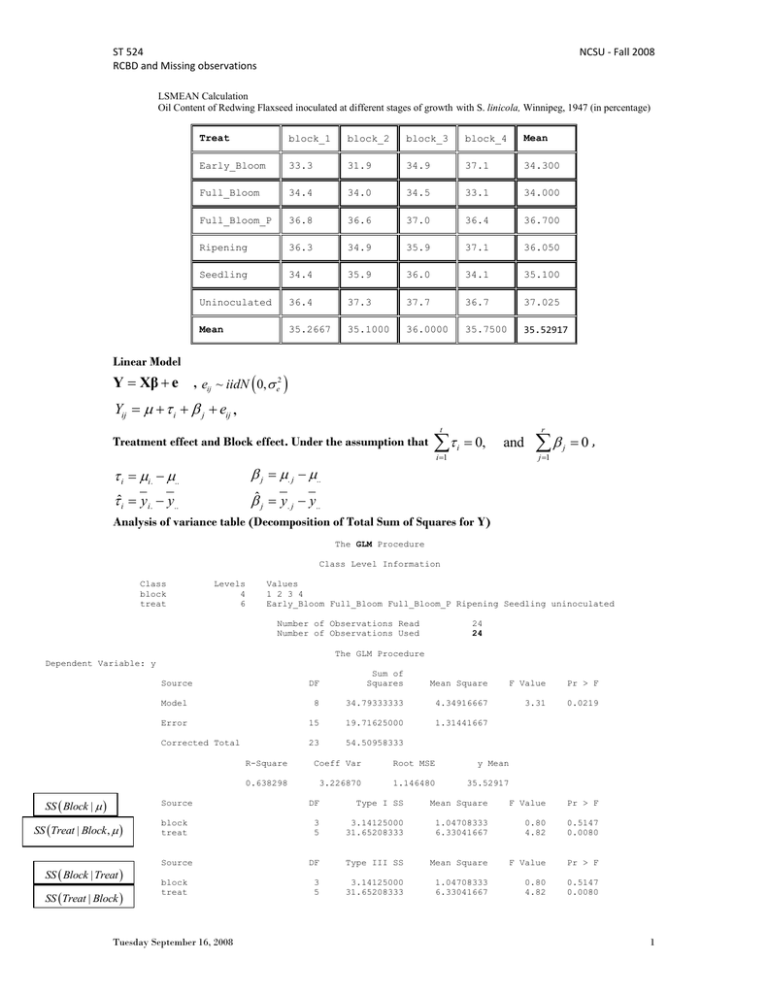
ST 524 RCBD and Missing observations NCSU - Fall 2008 LSMEAN Calculation Oil Content of Redwing Flaxseed inoculated at different stages of growth with S. linicola, Winnipeg, 1947 (in percentage) Treat block_1 block_2 block_3 block_4 Mean Early_Bloom 33.3 31.9 34.9 37.1 34.300 Full_Bloom 34.4 34.0 34.5 33.1 34.000 Full_Bloom_P 36.8 36.6 37.0 36.4 36.700 Ripening 36.3 34.9 35.9 37.1 36.050 Seedling 34.4 35.9 36.0 34.1 35.100 Uninoculated 36.4 37.3 37.7 36.7 37.025 Mean 35.2667 35.1000 36.0000 35.7500 35.52917 Linear Model Y Xβ e F , eij ~ iidN 0, e2 Yij i j eij , t Treatment effect and Block effect. Under the assumption that i 1 i i. .. j . j .. ˆi y i. y.. ˆ j y. j y.. i 0, r and j 1 j 0, Analysis of variance table (Decomposition of Total Sum of Squares for Y) The GLM Procedure Class Level Information Class block treat Levels 4 6 Values 1 2 3 4 Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated Number of Observations Read Number of Observations Used 24 24 The GLM Procedure Dependent Variable: y DF Sum of Squares Mean Square F Value Pr > F Model 8 34.79333333 4.34916667 3.31 0.0219 Error 15 19.71625000 1.31441667 Corrected Total 23 54.50958333 Source SS Block | Source SS Treat | Block , SS Block | Treat SS Treat | Block block treat Source block treat Tuesday September 16, 2008 R-Square Coeff Var Root MSE y Mean 0.638298 3.226870 1.146480 35.52917 DF Type I SS Mean Square F Value Pr > F 3 5 3.14125000 31.65208333 1.04708333 6.33041667 0.80 4.82 0.5147 0.0080 DF Type III SS Mean Square F Value Pr > F 3 5 3.14125000 31.65208333 1.04708333 6.33041667 0.80 4.82 0.5147 0.0080 1 ST 524 RCBD and Missing observations NCSU - Fall 2008 Balanced Design: Type I and Type III SS are the same: Block and Treatments are uncorrelated. Solution to linear model Parameter Intercept block block block block treat treat treat treat treat treat Predicted value for observations Standard Error t Value Pr > |t| 0.70207282 0.66192061 0.66192061 0.66192061 . 0.81068387 0.81068387 0.81068387 0.81068387 0.81068387 . 53.05 -0.73 -0.98 0.38 . -3.36 -3.73 -0.40 -1.20 -2.37 . <.0001 0.4765 0.3417 0.7110 . 0.0043 0.0020 0.6941 0.2477 0.0313 . Estimate 1 2 3 4 Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated 37.24583333 -0.48333333 -0.65000000 0.25000000 0.00000000 -2.72500000 -3.02500000 -0.32500000 -0.97500000 -1.92500000 0.00000000 B B B B B B B B B B B y11 , y23 , y64 37.2458 0.4833 0.6500 0.2500 yˆ11 1 1 0 0 0 1 0 0 0 0 0 0 37.2458 0.4833 2.725 34.0375 yˆ 1 0 0 1 0 0 1 0 0 0 0 2.725 37.2458 0.25 3.025 34.4708 23 yˆ 64 1 0 0 0 1 0 0 0 0 0 1 3.025 37.2458 37.2458 0.3250 0.9750 1.9250 0 Note that, predicted value is a function of the effects ˆi and ˆ j 34.0375 = 35.52917 + (34.3000 - 35.52917) + (35.2667 - 35.52917) 34.4708 = 35.52917 + (34.0000 - 35.52917) + (36.0000 - 35.52917) 37.2458 = 35.52917 + ( - 35.52917) + ( - 35.52917) Least Squares Mean for Treatment - Treatment LSMEAN Average for each treatment level over the block effects 37.2458 0.4833 0.6500 0.2500 ˆ1. 1 1 4 1 4 1 4 1 4 1 0 0 0 0 0 0 37.2458 1 4 0.4833 0.6500 0.2500 0 2.725 34.3 ˆ 1 1 4 1 4 1 4 1 4 0 1 0 0 0 0 2.725 37.2458 1 4 0.4833 0.6500 0.2500 0 3.025 34.0 2. 37.025 ˆ 6. 1 1 4 1 4 1 4 1 4 0 0 0 0 0 1 3.025 37.2458 1 4 0.4833 0.6500 0.2500 0 0 0.3250 0.9750 1.9250 0 Balanced Design with no missing observations: Simple Arithmetic Mean = Least Squares Mean ErrorMS 1.3144 LSMEAN Standard Error = 0.5732401 r 4 treat Standard Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated Tuesday September 16, 2008 y LSMEAN Error Pr > |t| 34.3000000 34.0000000 36.7000000 36.0500000 35.1000000 37.0250000 0.5732401 0.5732401 0.5732401 0.5732401 0.5732401 0.5732401 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 2 ST 524 RCBD and Missing observations NCSU - Fall 2008 Missing observations treat block_1 block_2 block_3 block_4 Mean Early_Bloom 33.3 31.9 34.9 37.1 34.300 Full_Bloom 34.4 . 34.5 33.1 34.000 Full_Bloom_P 36.8 36.6 37.0 36.4 36.700 Ripening 36.3 34.9 35.9 37.1 36.050 Seedling 34.4 35.9 36.0 34.1 35.100 Uninoculated 36.4 37.3 . 36.7 36.80 Mean 35.2667 35.3200 35.6600 35.7500 35.5000 The GLM Procedure Level of treat N Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated 4 3 4 4 4 3 Level of block N 1 2 3 4 6 5 5 6 --------------y-------------Mean Std Dev 34.3000000 34.0000000 36.7000000 36.0500000 35.1000000 36.8000000 2.23308456 0.78102497 0.25819889 0.91469485 0.98994949 0.45825757 --------------y-------------Mean Std Dev 35.2666667 35.3200000 35.6600000 35.7500000 1.41938954 2.10760528 0.98640762 1.71551741 Analysis of Variance Table The GLM Procedure Dependent Variable: y Sum of DF Source SS Treat | Block , SS Block | Treat , SS Treat | Block , Mean Square F Value Pr > F 2.35 0.0820 Model 8 28.06797619 3.50849702 Error 13 19.37202381 1.49015568 Corrected Total 21 47.44000000 R-Square Coeff Var Root MSE y Mean 0.591652 3.438646 1.220719 35.50000 Source SS Block | Squares block treat Source block treat Missing observations: Tuesday September 16, 2008 Type I SS DF Type I SS Mean Square F Value Pr > F 3 5 0.99166667 27.07630952 0.33055556 5.41526190 0.22 3.63 0.8795 0.0282 DF Type III SS Mean Square F Value Pr > F 3 5 2.87797619 27.07630952 0.95932540 5.41526190 0.64 3.63 0.6005 0.0282 Type III SS 3 ST 524 RCBD and Missing observations NCSU - Fall 2008 Solution Parameter Intercept block block block block treat treat treat treat treat treat Estimate 37.21488095 -0.48333333 -0.76130952 0.20297619 0.00000000 -2.65446429 -3.12142857 -0.25446429 -0.90446429 -1.85446429 0.00000000 1 2 3 4 Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated B B B B B B B B B B B Standard Error t Value Pr > |t| 0.81834165 0.70478263 0.75049541 0.75049541 . 0.94591683 1.03169613 0.94591683 0.94591683 0.94591683 . 45.48 -0.69 -1.01 0.27 . -2.81 -3.03 -0.27 -0.96 -1.96 . <.0001 0.5049 0.3289 0.7911 . 0.0149 0.0097 0.7921 0.3564 0.0717 . Least Squares Mean for Treatment - Treatment LSMEAN Average for each treatment level over the block effects 37.2149 2.6545 3.1214 0.2545 ˆ1. 1 1 0 0 0 0 0 1 4 1 4 1 4 1 4 0.9045 37.2149 2.6545 1 4 0.4833 0.7613 0.2030 0 34.3 ˆ 1 0 1 0 0 0 0 1 4 1 4 1 4 1 4 1.8545 37.2149 3.1214 1 4 0.4833 0.7613 0.2030 0 33.8331 2. 36.9545 ˆ 6. 1 0 0 0 0 0 1 1 4 1 4 1 4 1 4 0 37.2149 0 1 4 0.4833 0.7613 0.2030 0 0.4833 0.7613 0.2030 0 37.2149 0.4833 0.7613 0.2030 ˆ1. 1 1 4 1 4 1 4 1 4 1 0 0 0 0 0 0 37.2149 1 4 0.4833 0.761 3 0.2030 0 2.6545 34.3 ˆ 1 1 4 1 4 1 4 1 4 0 1 0 0 0 0 2.6545 37.2149 1 4 0.4833 0.7613 0.2030 0 3.1214 33.8331 2. 36.9545 ˆ 6. 1 1 4 1 4 1 4 1 4 0 0 0 0 0 1 3.1214 37.2149 1 4 0.4833 0.7613 0.2030 0 0 0.2545 0.9045 1.8545 0 Standard treat y LSMEAN Error Pr > |t| Early_Bloom Full_Bloom Full_Bloom_P Ripening Seedling uninoculated Balanced Design with missing observations: Least squares means are estimates of 34.3000000 33.8330357 36.7000000 36.0500000 35.1000000 36.9544643 Simple Arithmetic Mean i 0.6103597 0.7226477 0.6103597 0.6103597 0.6103597 0.7226477 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 Least Squares Mean ˆ1* ˆ2* ˆ3* ˆ4* * * * ˆ ˆi ˆ 4 ˆ i. ˆ * ˆi* Pairwise differences of LSMEANS are free of block effects. ErrorMS 1.49015568 0.6103597, for r 4, ri ri but, se(Full Bloom LSMEAN) = 0.7226 0.7048 LSMEAN Standard Error = Tuesday September 16, 2008 0.7047826, for r 3 4 ST 524 RCBD and Missing observations NCSU - Fall 2008 To calculate LSMEANS we can use the ESTIMATE statement in PROC GLM or proc MIXED Label LSMEAN LSMEAN LSMEAN LSMEAN LSMEAN LSMEAN T1 T2 T3 T4 T5 T6 For LSMEAN (Full BLOOM) estimate "LSMEAN T2" intercept 4 Estimate Estimates Standard Error DF t Value Pr > |t| 34.3000 33.8330 36.7000 36.0500 35.1000 36.9545 0.6104 0.7226 0.6104 0.6104 0.6104 0.7226 13 13 13 13 13 13 56.20 46.82 60.13 59.06 57.51 51.14 <.0001 <.0001 <.0001 <.0001 <.0001 <.0001 treat 0 4 block 1 1 1 1/divisor=4; Simple Mean differences includes block effects y Early Bloom y Full Bloom y11 y12 y13 y14 y21 y23 y24 4 3 1 1 e11 1 2 e12 1 3 e13 1 4 e14 2 1 e21 2 3 e23 1 4 e24 3 4 e e e e e e e 3 4 4 1 1 2 11 12 13 14 2 1 3 21 23 24 4 4 3 3 1 3 2 3 4 1 2 e1. e2. 12 LSMEANS differences is free of block effects ˆ1* 2 ˆ2* ˆ3* ˆ4* ˆ * ˆ2* ˆ3* ˆ4* * * Y Xβ e ˆ e , eij ˆ~* iidN ˆ* 0, ˆ * ˆ2* 1 ˆ Early ˆ1 ˆ2 Bloom Full Bloom 1 4 L ' X ' X MS Error L L ' 1 1 4 1 4 1 4 1 4 0 1 0 0 0 0 Var(LSMEAN for Full Bloom) = X 'X 0.375 -0.166667 -0.166667 -0.166667 0 -0.25 -0.25 -0.25 -0.25 -0.25 0 4 where, for Treat = 2, = -0.166667 0.3333333 0.1666667 0.1666667 0 0 0 0 0 0 0 -0.166667 0.1666667 0.3333333 0.1666667 0 0 0 0 0 0 0 -0.166667 0.1666667 0.1666667 0.3333333 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 0 -0.25 0 0 0 0 0.5 0.25 0.25 0.25 0.25 0 -0.25 0 0 0 0 0.25 0.5 0.25 0.25 0.25 0 -0.25 0 0 0 0 0.25 0.25 0.5 0.25 0.25 0 -0.25 0 0 0 0 0.25 0.25 0.25 0.5 0.25 0 -0.25 0 0 0 0 0.25 0.25 0.25 0.25 0.5 0 0 0 0 0 0 0 0 0 0 0 0 SAS MANUAL (PROC GLM) LS-means are, in effect, within-group means appropriately adjusted for the other effects in the model. More precisely, they estimate the marginal means for a balanced population (as opposed to the unbalanced design). For this reason, they are also called estimated population marginal means (Searle, 1980). In the same way that the Type I F-test assesses differences between the arithmetic treatment means (when the treatment effect comes first in the model), the Type III F-test assesses differences between the LS-means. Accordingly, for the unbalanced twoway design, the discrepancy between the Type I and Type III tests is reflected in the arithmetic treatment means and treatment LS-means. Note that, while the arithmetic means are always uncorrelated (under the usual assumptions for analysis of variance), the LS-means may not be. This fact complicates the problem of multiple comparisons for LS-means. Estimable functions Type I hypothesis Tuesday September 16, 2008 5 ST 524 RCBD and Missing observations NCSU - Fall 2008 Sum to zero parameterization Last level is 0 parameterization, default parameterization in sas Tuesday September 16, 2008 6 ST 524 RCBD and Missing observations NCSU - Fall 2008 Randomized Complete Block Design with Subsampling Example Data from a study in potato Late Blight resistance conducted by researcher Luis Rivera at the International Potato Center in Lima, Peru n 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 family fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 Block Yield n 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 1 150 50 300 350 650 500 450 500 250 350 150 150 150 100 150 300 100 100 300 60 500 100 250 350 500 300 400 350 400 250 1300 100 350 200 200 300 100 300 300 80 150 500 500 200 270 220 900 200 340 900 390 530 120 50 150 150 420 490 140 300 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 Tuesday September 16, 2008 family fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 Block 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 2 Yield n 700 900 300 150 450 700 350 400 200 350 300 500 50 300 50 300 200 750 550 300 60 70 60 110 150 10 190 150 140 150 350 100 100 250 100 100 150 500 650 200 200 620 300 130 240 100 250 230 300 150 400 500 250 900 750 50 400 250 100 400 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 1 2 3 4 5 6 7 8 9 10 family fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam01 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam24 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam25 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam31 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam32 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 fam34 Block Yield 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 3 400 150 250 100 750 250 750 200 250 800 20 370 400 100 300 300 600 80 250 200 230 140 370 30 20 320 120 130 410 350 320 20 110 280 160 110 60 80 120 100 250 400 180 220 120 250 450 250 450 130 600 200 150 300 300 300 350 650 200 450 7 ST 524 RCBD and Missing observations NCSU - Fall 2008 Linear Model : Yij i j ij ijk t Treatment is a Fixed Effect i 1 0 i r Block is a Fixed Effect j 1 j 0 Plot is a Random effect , nested on treatments Plant is a Random effect, nested on plots ij and ijk ij ~ iidN 0, 2 ijk ~ iidN 0, 2 , are independent random effects Analysis of Variance - Expected Mean Squares Sum of Squares Decomposition t r s i 1 j 1 k 1 t r t i 1 j 1 r s (Yijk Y ... )2 rs (Y i.. Y ... )2 ts (Y . j. Y ... )2 s (Y ij. Y i.. Y . j. )2 (Yijk Y ij. )2 i 1 j 1 i j k 1 SS due to Treatment Effects SS due to Block Effects SS due to Sampling Error Variation among samples within pots SS due to Experimental Error Variation among pots within treatments Analysis of Variance Table Sources df Sum of Squares r Block r-1 ts (Y . j. Y ... ) 2 j 1 t Treatment t-1 rs (Y i.. Y ... ) 2 i 1 Experimental Error Sampling Error t 1 r 1 r i 1 j 1 t r s 1 Plants(Plot) Corrected Total t s (Y ij . Y i.. Y . j. )2 t r s 1 s (Y i j k 1 t r s i 1 j 1 k 1 Tuesday September 16, 2008 ijk Y ij . )2 Mean Square Block SS r 1 E(MS) 2 s e2 t s Treatment SS t 1 2 s e2 r s Exp Error SS r 1 t 1 2 s 2 Sampling Error SS rt s 1 2 2 j j r 1 2 i i t 1 (Yijk Y ... )2 8 ST 524 RCBD and Missing observations NCSU - Fall 2008 SAS Output Dependent Variable: Yield Source DF Sum of Squares Mean Square F Value Pr > F Model 17 1619185.000 95246.176 2.43 0.0021 Error 162 6351310.000 39205.617 Corrected Total 179 7970495.000 R-Square Coeff Var Root MSE Yield Mean 0.203147 67.53977 198.0041 293.1667 Source DF Type I SS Mean Square F Value Pr > F Block 2 52990.0000 26495.0000 0.68 0.5102 family 5 727285.0000 145457.0000 3.71 0.0033 family*Block 10 838910.0000 83891.0000 2.14 0.0242 Source DF Type III SS Mean Square F Value Pr > F Block 2 52990.0000 26495.0000 0.68 0.5102 family 5 727285.0000 145457.0000 3.71 0.0033 family*Block 10 838910.0000 83891.0000 2.14 0.0242 Expected Mean Squares table The GLM Procedure Source Type III Expected Mean Square Block Var(Error) + 10 Var(family*Block) + Q(Block) family Var(Error) + 10 Var(family*Block) + Q(family) family*Block Var(Error) + 10 Var(family*Block) Test of Hypothesis Source DF Type III SS Mean Square F Value Pr > F Block 2 52990 26495 0.32 0.7362 family 5 727285 145457 1.73 0.2145 Error 10 838910 83891 Error: MS(family*Block) Tuesday September 16, 2008 9 ST 524 RCBD and Missing observations NCSU - Fall 2008 Source DF Type III SS Mean Square F Value Pr > F family*Block 10 838910 83891 2.14 0.0242 162 6351310 39206 Error: MS(Error) Treatments H o : 1 2 3 4 5 6 0 H1 : At least one i 0 , i 1, 2, Fcalc 145457.0 1.73 83891.0 ,6 p-value =0.2145 Do not Reject Ho Experimental error H o : 2 0 H1 : 2 0 Fcalc 83891 2.14 39206 p-value = 0.0242 Reject Ho Variance Components Estimation Var(among subsamples units within plot and family): 2 Var(among experimental units): 2 =39206 83891 39206 4468.5 10 Variance among plants within plots Variance among experimental units An Observation, Yij i j ij ijk Variance of an observation Var Yijk 2 2 estimated by 4468.5 + 39206 = 43674.5 Plot mean Y ij. i j ij ijk k s i j ij ij. s Variance of a plot mean 2 , estimated by 4468.5 39206 8389.1 s 10 Note that estimate value for the variance of a plot mean is given by Experimental Error MS 83891 var Y ij . 8389.1 s 10 Var Y ij . Var i j ij ij . 2 Treatment mean j Y i.. i jk rs jk rs ij ijk jk rs Variance of a treatment mean Tuesday September 16, 2008 10 ST 524 RCBD and Missing observations Var Y i.. NCSU - Fall 2008 j j 2 2 4468.5 39206 2796.3667 Var i i. i.. , estimated by 3 3 10 r sr r Note that the estimated value of variance of a treatment mean is given by Experimental Error MS 83891 var Y i.. 2796.3667 rs 3 10 Var Y i... 2796.6667 52.8807 Standard error of a treatment mean = PROC GLM Least Squares Means Standard Errors and Probabilities Calculated Using the Type III MS for family*Block as an Error Term family Yield LSMEAN Standard Error Pr > |t| fam01 398.333333 52.880683 <.0001 fam24 249.333333 52.880683 0.0008 fam25 220.333333 52.880683 0.0019 fam31 236.333333 52.880683 0.0012 fam32 313.333333 52.880683 0.0001 fam34 341.333333 52.880683 <.0001 PROC MIXED OUTPUT Covariance Parameter Estimates Cov Parm Subject Estimate Intercept family*Block Residual 4468.54 39206 Wald’s test for Covariance paramters Asymptotically Normal Wald’s Z test Covariance Parameter Estimates Cov Parm Subject Intercept family*Block Residual Estimate Standard Error Z Value Pr Z 4468.54 3776.93 1.18 0.1184 39206 4356.18 9.00 <.0001 H o : 2 0 H1 : 2 0 H o : 2 0 H1 : 2 0 Type 3 Tests of Fixed Effects Tuesday September 16, 2008 Effect Num DF Den DF F Value Pr > F Block 2 10 0.32 0.7362 family 5 10 1.73 0.2145 11 ST 524 RCBD and Missing observations NCSU - Fall 2008 Least Squares Means Effect family Estimate Standard Error DF t Value Pr > |t| family fam01 398.33 52.8807 10 7.53 <.0001 family fam24 249.33 52.8807 10 4.72 0.0008 family fam25 220.33 52.8807 10 4.17 0.0019 family fam31 236.33 52.8807 10 4.47 0.0012 family fam32 313.33 52.8807 10 5.93 0.0001 family fam34 341.33 52.8807 10 6.45 <.0001 Same results when working with average over plants within each plot Analyzing Block*Family interaction Test of hypothesis for family is valid when no block*family interaction is present, families should behave similarly within each block. Tuesday September 16, 2008 12 ST 524 RCBD and Missing observations Tuesday September 16, 2008 NCSU - Fall 2008 13