Interaction of N-terminal Peptides of Glycogen Phosphorylase with Calmodulin
advertisement

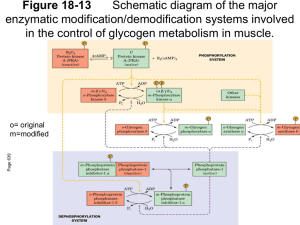
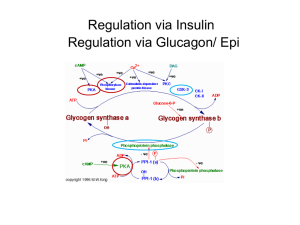
Interaction of N-terminal Peptides of Glycogen Phosphorylase with Calmodulin By James Proestos Dr. Sonia Anderson’s Lab Biochemistry and Biophysics Department Glycogen Phosphorylase Information • Found in fast twitch muscle tissue • It catalyzes the breakdown of glycogen • Controlled by phosphorylation/ dephosphorylation The Phosphorylated and Unphosphorylated States of Glycogen Phosphorylase phosphorylase b phosphorylase a Substrate(s) Serine 14 Calmodulin Structure • Is found in all animal and plant tissues • Binding of calcium controls its ability to bind to a protein to regulate the target protein’s activity Calmodulin Binding Process 2+ 4 Ca Protein Cascade of Reactions in Glycogen Degradation Hormonal and Calcium control The Interaction of Proteins in Glycogen Cascade • Phosphorylase kinase becomes active by calcium binding to the intrinsic calmodulin • The phosphorylase kinase interacts with the glycogen phosphorylase • It is not known if the calmodulin can readily bind with glycogen phosphorylase in this interaction Calmodulin/Phosphorylase B Interaction Rabbit Muscle Extract Bound Calmodulin/ Sepharose gel Peptides that do not bind to calmodulin SDS Page of Rabbit Muscle Extract 96 K 68 K 42 K 29 K 18 K 12 K Hypothesis • Malencik and Anderson proposed that calmodulin binding regions are often sites of regulation by serinethreonine phosphorylation/dephosphorylation Hypothesis • Malencik and Anderson proposed that calmodulin binding regions are often sites of regulation by serinethreonine phosphorylation/dephosphorylation Question • Is the calmodulin binding region of phosphorylase b the same as the phosphorylation site and how does phosphorylation affect this binding to calmodulin? Phosphorylase Purification Ammonium Sulfate Precipitation and Selective Crystallization Purification of Calmodulin SDS Page of stages in calmodulin purification • Four column chromatographies; 3000 fold purification • 96 K 68 K 42 K 29 K 18 K 12 K Cleavage of Phosphorylase B 1 14 841 Subtilisin 1 14 Hydroxylamine CNBR RXN 264 1 1 265 14 14 134 91 841 135 242 350 351 428 442 604 259 260 497 498 841 Cleavage of Phosphorylase B 1 14 841 CNBR RXN 1 14 91 242 350 351 428 442 604 Peptide 1-91 Purification Cation Exchange Synthetic Peptide 5-20 5 14 SNQQLKRQISVRGLAG 20 Synthetic Peptide 5-20 5 14 20 SNQQLKRQISVRGLAG -P +P Analysis of Calmodulin/Glycogen Phosphorylase Interaction Determine Affinity of calmodulin-peptide complex by the use of dansyl calmodulin fluorescence Isolated peptides A)Peptide(1-91) B)Peptide(5-20) C)CaM Binding Peptide(s) Phosphorylate peptides and recheck affinity Fluorescence Titration Analysis of Peptide-Calmodulin Interactions Peptide -P Affinity +P 1-91 40 nM 60 nM 5-20 93 uM 225 uM Selenoprotein W 18 nM -- (KFRKLVTAIKAALAQ) Melittin <1 nM -- Conclusion • The N-terminal peptide(5-20) of phosphorylase binds to calmodulin • Phosphorylation of this peptide weakens the interaction with calmodulin • Peptide 1-91 binds more tightly to calmodulin than does peptide(5-20) (µM vs. nM) • The affinity of peptide 1-91 compared to 5-20 suggests that additional sequences in phosphorylase participate in calmodulin binding SRPLSDQEKRKQISVRGLAGVENVTELKKNFNRHLHFTLVKDRNVATPRDYYFALAHT VRDHLVGRWIRTQQHYYEKDPKRIYYLSLEFYM Acknowledgement Howard Hughes Medical Institute Dr. Sonia Anderson Dean Malencik Andy Bauman Department of Biochemistry and Biophysics Kevin Ahern

![Anti-Calmodulin antibody [J4D8] ab75207 Product datasheet 1 References 1 Image](http://s2.studylib.net/store/data/012961983_1-fbc3a125831a6d5a42cfe8d3e7d70ccc-300x300.png)