Y. Zhang , E.M. Smith , T. Mersha , J. Gunnell
advertisement
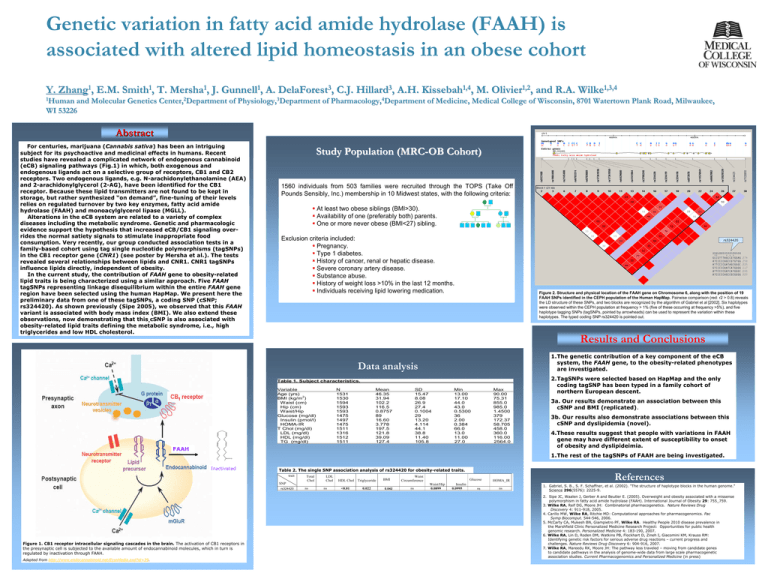
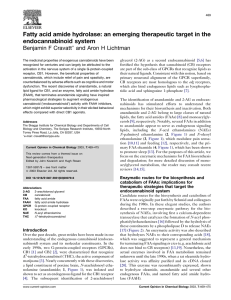
Y. 1 Zhang , E.M. 1 Smith , T. 1 Mersha , J. 1 Gunnell , A. 3 DelaForest , C.J. 3 Hillard , A.H. 1,4 Kissebah , M. 1,2 Olivier , and R.A. 1,3,4 Wilke 1Human and Molecular Genetics Center,2Department of Physiology,3Department of Pharmacology,4Department of Medicine, Medical College of Wisconsin, 8701 Watertown Plank Road, Milwaukee, WI 53226 Abstract For centuries, marijuana (Cannabis sativa) has been an intriguing subject for its psychoactive and medicinal effects in humans. Recent studies have revealed a complicated network of endogenous cannabinoid (eCB) signaling pathways (Fig.1) in which, both exogenous and endogenous ligands act on a selective group of receptors, CB1 and CB2 receptors. Two endogenous ligands, e.g. N-arachidonylethanolamine (AEA) and 2-arachidonylglycerol (2-AG), have been identified for the CB1 receptor. Because these lipid transmitters are not found to be kept in storage, but rather synthesized “on demand”, fine-tuning of their levels relies on regulated turnover by two key enzymes, fatty acid amide hydrolase (FAAH) and monoacylglycerol lipase (MGLL). Alterations in the eCB system are related to a variety of complex diseases including the metabolic syndrome. Genetic and pharmacologic evidence support the hypothesis that increased eCB/CB1 signaling overrides the normal satiety signals to stimulate inappropriate food consumption. Very recently, our group conducted association tests in a family-based cohort using tag single nucleotide polymorphisms (tagSNPs) in the CB1 receptor gene (CNR1) (see poster by Mersha et al.). The tests revealed several relationships between lipids and CNR1. CNR1 tagSNPs influence lipids directly, independent of obesity. In the current study, the contribution of FAAH gene to obesity-related lipid traits is being characterized using a similar approach. Five FAAH tagSNPs representing linkage disequilibrium within the entire FAAH gene region have been selected using the human HapMap. We present here the preliminary data from one of these tagSNPs, a coding SNP (cSNP; rs324420). As shown previously (Sipe 2005), we observed that this FAAH variant is associated with body mass index (BMI). We also extend these observations, now demonstrating that this cSNP is also associated with obesity-related lipid traits defining the metabolic syndrome, i.e., high triglycerides and low HDL cholesterol. Study Population (MRC-OB Cohort) 1560 individuals from 503 families were recruited through the TOPS (Take Off Pounds Sensibly, Inc.) membership in 10 Midwest states, with the following criteria: At least two obese siblings (BMI>30). Availability of one (preferably both) parents. One or more never obese (BMI<27) sibling. Exclusion criteria included: Pregnancy. Type 1 diabetes. History of cancer, renal or hepatic disease. Severe coronary artery disease. Substance abuse. History of weight loss >10% in the last 12 months. Individuals receiving lipid lowering medication. rs324420 Figure 2. Structure and physical location of the FAAH gene on Chromosome 6, along with the position of 19 FAAH SNPs identified in the CEPH population of the Human HapMap. Pairwise comparison (red: r2 > 0.8) reveals the LD structure of these SNPs, and two blocks are recognized by the algorithm of Gabriel et al [2002]. Six haplotypes were observed within the CEPH population at frequency > 1% (five of these occurring at frequency >5%), and five haplotype tagging SNPs (tagSNPs, pointed by arrowheads) can be used to represent the variation within these haplotypes. The typed coding SNP rs324420 is pointed out. Results and Conclusions 1.The genetic contribution of a key component of the eCB system, the FAAH gene, to the obesity-related phenotypes are investigated. Data analysis Table 1. Subject characteristics. Variable Age (yrs) 2 BMI (kg/m ) Waist (cm) Hip (cm) Waist/Hip Glucose (mg/dl) Insulin (pmol/l) HOMA-IR T Chol (mg/dl) LDL (mg/dl) HDL (mg/dl) TG (mg/dl) N 1531 1530 1594 1593 1593 1475 1497 1475 1511 1316 1512 1511 Mean 46.35 31.94 102.2 116.5 0.8757 89 16.60 3.778 197.5 121.8 39.09 127.4 SD 15.47 8.08 26.9 27.4 0.1004 29 13.20 4.114 44.1 38.8 11.40 105.8 Min 13.00 17.10 44.0 43.0 0.5300 36 2.00 0.384 66.0 13.0 11.00 27.0 Max 90.00 75.31 855.0 985.0 1.4500 379 172.37 58.705 458.0 360.0 116.00 2564.0 FAAH 3a. Our results demonstrate an association between this cSNP and BMI (replicated). 3b. Our results also demonstrate associations between this cSNP and dyslipidemia (novel). 4.These results suggest that people with variations in FAAH gene may have different extent of susceptibility to onset of obesity and dyslipideimia. 1.The rest of the tagSNPs of FAAH are being investigated. Inactivated Table 2. The single SNP association analysis of rs324420 for obesity-related traits. trait SNP rs324420 Figure 1. CB1 receptor intracellular signaling cascades in the brain. The activation of CB1 receptors in the presynaptic cell is subjected to the available amount of endocannabinoid molecules, which in turn is regulated by inactivation through FAAH. Adapted from http://www.endocannabinoid.net/EcsnMedia.axd?id=39. 2.TagSNPs were selected based on HapMap and the only coding tagSNP has been typed in a family cohort of northern European descent. Total Chol ns LDL Chol ns HDL Chol <0.01 Triglyceride 0.022 BMI 0.042 Waist Circumference ns Glucose Waist/Hip 0.0899 Insulin 0.0995 ns HOMA_IR ns References 1. Gabriel, S. B., S. F. Schaffner, et al. (2002). "The structure of haplotype blocks in the human genome." Science 296(5576): 2225-9. 2. Sipe JC, Waalen J, Gerber A and Beutler E. (2005). Overweight and obesity associated with a missense polymorphism in fatty acid amide hydrolase (FAAH). International Journal of Obesity 29: 755_759. 3. Wilke RA, Reif DG, Moore JH: Combinatorial pharmacogenetics. Nature Reviews Drug Discovery 4: 911-918, 2005. 4. Carillo MW, Wilke RA, Ritchie MD: Computational approaches for pharmacogenomics. Pac Symp Biocomput. 544-546, 2006. 5. McCarty CA, Mukesh BN, Giampietro PF, Wilke RA. Healthy People 2010 disease prevalence in the Marshfield Clinic Personalized Medicine Research Project: Opportunities for public health genomic research. Personalized Medicine 4: 183-190, 2007. 6. Wilke RA, Lin D, Roden DM, Watkins PB, Flockhart D, Zineh I, Giacomini KM, Krauss RM: Identifying genetic risk factors for serious adverse drug reactions – current progress and challenges. Nature Reviews Drug Discovery 6: 904-916, 2007. 7. Wilke RA, Mareedu RK, Moore JH: The pathway less traveled – moving from candidate genes to candidate pathways in the analysis of genome-wide data from large scale pharmacogenetic association studies. Current Pharmacogenomics and Personalized Medicine (in press)