C ommentary
advertisement
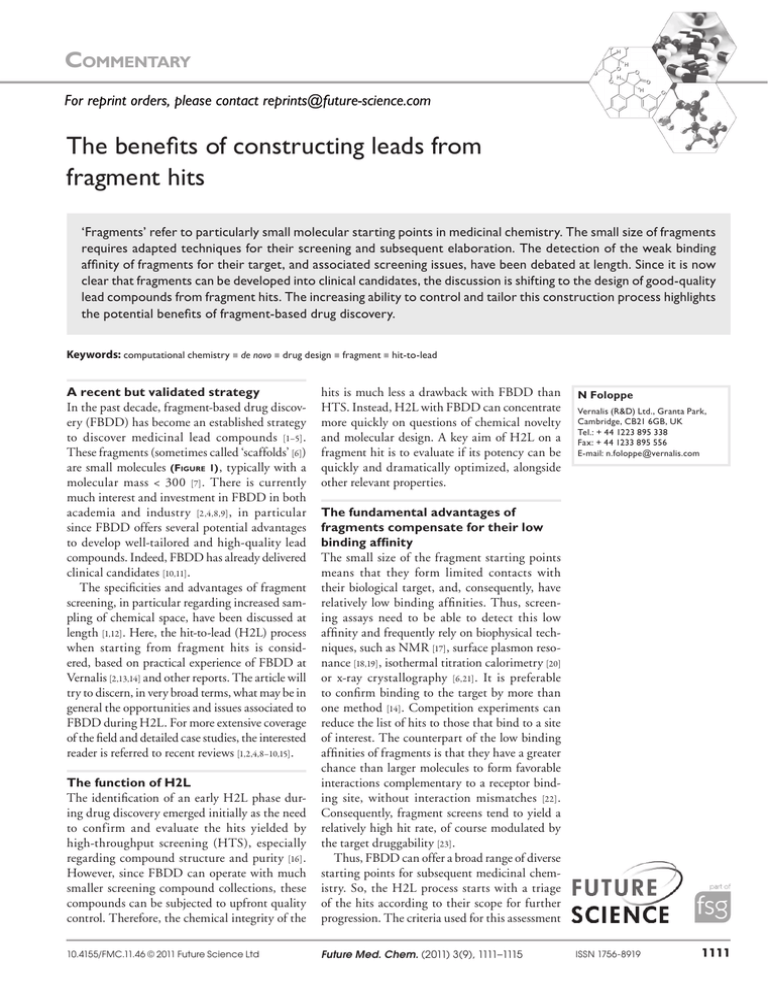
Commentary For reprint orders, please contact reprints@future-science.com The benefits of constructing leads from fragment hits ‘Fragments’ refer to particularly small molecular starting points in medicinal chemistry. The small size of fragments requires adapted techniques for their screening and subsequent elaboration. The detection of the weak binding affinity of fragments for their target, and associated screening issues, have been debated at length. Since it is now clear that fragments can be developed into clinical candidates, the discussion is shifting to the design of good-quality lead compounds from fragment hits. The increasing ability to control and tailor this construction process highlights the potential benefits of fragment-based drug discovery. Keywords: computational chemistry n de novo n drug design n fragment n hit-to-lead A recent but validated strategy In the past decade, fragment-based drug discovery (FBDD) has become an established strategy to discover medicinal lead compounds [1–5] . These fragments (sometimes called ‘scaffolds’ [6]) are small molecules (Figure 1) , typically with a molecular mass < 300 [7] . There is currently much interest and investment in FBDD in both academia and industry [2,4,8,9] , in particular since FBDD offers several potential advantages to develop well-tailored and high-quality lead compounds. Indeed, FBDD has already delivered clinical candidates [10,11] . The specificities and advantages of fragment screening, in particular regarding increased sampling of chemical space, have been discussed at length [1,12] . Here, the hit-to-lead (H2L) process when starting from fragment hits is considered, based on practical experience of FBDD at Vernalis [2,13,14] and other reports. The article will try to discern, in very broad terms, what may be in general the opportunities and issues associated to FBDD during H2L. For more extensive coverage of the field and detailed case studies, the interested reader is referred to recent reviews [1,2,4,8–10,15] . hits is much less a drawback with FBDD than HTS. Instead, H2L with FBDD can concentrate more quickly on questions of chemical novelty and molecular design. A key aim of H2L on a fragment hit is to evaluate if its potency can be quickly and dramatically optimized, alongside other relevant properties. The function of H2L The identification of an early H2L phase during drug discovery emerged initially as the need to confirm and evaluate the hits yielded by high-throughput screening (HTS), especially regarding compound structure and purity [16] . However, since FBDD can operate with much smaller screening compound collections, these compounds can be subjected to upfront quality control. Therefore, the chemical integrity of the The fundamental advantages of fragments compensate for their low binding affinity The small size of the fragment starting points means that they form limited contacts with their biological target, and, consequently, have relatively low binding affinities. Thus, screening assays need to be able to detect this low affinity and frequently rely on biophysical techniques, such as NMR [17] , surface plasmon resonance [18,19] , isothermal titration calorimetry [20] or x-ray crystallography [6,21] . It is preferable to confirm binding to the target by more than one method [14] . Competition experiments can reduce the list of hits to those that bind to a site of interest. The counterpart of the low binding affinities of fragments is that they have a greater chance than larger molecules to form favorable interactions complementary to a receptor binding site, without interaction mismatches [22] . Consequently, fragment screens tend to yield a relatively high hit rate, of course modulated by the target druggability [23] . Thus, FBDD can offer a broad range of diverse starting points for subsequent medicinal chemistry. So, the H2L process starts with a triage of the hits according to their scope for further progression. The criteria used for this assessment 10.4155/FMC.11.46 © 2011 Future Science Ltd Future Med. Chem. (2011) 3(9), 1111–1115 N Foloppe Vernalis (R&D) Ltd., Granta Park, Cambridge, CB21 6GB, UK Tel.: + 44 1223 895 338 Fax: + 44 1233 895 556 E-mail: n.foloppe@vernalis.com ISSN 1756-8919 1111 Commentary | Foloppe OH N HO O O O H N N N CO2Me O NH2 N N NH2 NH2 N H2N N H S N N Fragment bound to Hsp90 (x-ray structure) OH Asp93 Elaborate fragment (hit-to-lead) HO N O N NHEt O O N-AUY922/VER-52296VP Clinical candidate in Phase II (oncology) Figure 1. Schematic illustration of the potential of fragments for their development in clinical candidates. The top section shows examples of fragments, which were found to bind to the ATP-binding site of the Hsp90 protein (an oncology target) at Vernalis R&D Ltd [44] . These small molecules are representative of the small size of fragments. Accordingly, these compounds had weak binding affinities for Hsp90. However, visualization of the fragment-binding modes to the protein, combined with molecular modeling and medicinal chemistry, led to the progress of a fragment hit into a potent lead compound, which is now in phase II clinical trials. Hsp90: Heat shock protein 90. include chemical novelty, binding affinity and ligand efficiency (LE) [24] , whether the binding mode to the target can be visualized, amenability to synthetic chemistry, and, sometimes, clues of a functional profile (e.g., agonist or antagonist [11]). The weighting of these criteria is, of course, context dependent. The selected fragment is intuitively seen as the core of its derived chemical series, therefore, its novelty affects the patentability of derived analogs, especially with target classes for which an intellectual property niche is at a premium. The LE for such a core is another important parameter, since a high initial efficiency indicates that the starting fragment makes very favorable interactions with the target, normalized with respect to its number of nonhydrogen atoms. The LE of the initial fragment influences the LE achieved by derived analogs [25] . A high starting LE is preferred because it signals the opportunity to obtain high-affinity lead compounds without adding undue molecular mass, offering a promising start towards oral bioavailability of the derived lead [26] . 1112 Future Med. Chem. (2011) 3(9) Accelerating H2L with structural information Since most fragments hits have low binding affinities, a first challenge is to increase their affinity substantially, on a pragmatic timescale. This process is greatly facilitated in view of the fragmentbinding mode to its target, usually ascertained by x-ray crystallography [6,11,13,15,19,21,27–30] . The structure of the bound fragment accelerates the H2L process since it helps to prioritize among an otherwise very large number of elaboration options, including those with respect to target selectivity [11,30] . Such structure-guided elaboration assumes that the initial binding mode of the fragment is robust and not prone to changes upon derivatization. This requirement is frequently met, especially when the fragments forms asymmetrical polar interactions with the receptor. Yet, counter examples have been described [19,28,31,32] . One may argue that alteration of a binding mode with different substituents only adds to the range of options for H2L progression. In our experience, binding mode switches do not typically future science group The benefits of constructing leads from fragments hits hamper progress, confirming that FBDD lends itself to a controlled constructive approach for the design of lead compounds. The structure-based construction of leads from ideally positioned starting points gives the opportunity to tailor those leads for the target from the outset of H2L. Monitoring changes in LE at each step helps select the most efficient substituents for binding, to increase binding affinity while keeping the compound size under control. Smaller compounds tend to have more favorable absorption and distribution properties [26] . Other relevant physicochemical properties, as well as the selectivity profile, can be monitored and influenced incrementally when building from fragment to lead. Indeed a small (unencumbered) starting point offers, in principle, the opportunity to follow an optimal path from hit to lead. The fragment-binding mode gives early indications regarding which bond vectors will be engaged in optimization of binding affinity, and which vectors are amenable to more diverse variations to tune overall physicochemical properties. Thus, H2L with FBDD contrasts with situations where one is faced with a relatively large hit with mediocre LE, where much effort may be spent redesigning the starting point before productive optimization can be tackled. With HTS hits, an objective of H2L can be to find the minimum active fragment in a complex molecule [16] . Instead, FBDD starts immediately from a suitable core with a promising binding mode and can concentrate more directly on the path to a lead with well-balanced physicochemical properties for druglikeness [4] . Explore & design with computational chemistry Seeing how a fragment binds to its target gives a helpful first impression. However, to be most productive, FBDD can exploit the full range of drug-design tools available from modern computational chemistry. When applicable, one spontaneously overlays and visualizes the bound fragment with other, larger, compounds binding to the same site. Larger compounds will typically extend to regions of the binding site not occupied by the fragment; hence, one naturally tries to transfer elements from the larger compound to substitute the smaller fragment. For this to work, the exchanged covalent bonds need to be well aligned in a precise local overlay, and the associated intramolecular effects must also be compatible. These constraints make merging between compounds more difficult than future science group | Commentary it appears, but there are examples of hybrid compounds evolved from a fragment [9,13] . The combinatorial hybridization of overlaid ligands has been implemented in the BREED algorithm [33] . It assumes that the binding mode of several (many) potent compounds is available; computational docking can help obtain this information quickly for elaborated compounds reported in the literature, without always having to synthesize and crystallize them. Another tactic is to enumerate virtual libraries around the fragment core, according to predefined chosen chemistries and with accessible reagents. Then, the virtual library can be docked to the binding site, while enforcing the experimentally determined binding mode of the fragment core [6,11] . This explores the conformations of the putative substituents around the fragment core and their molecular recognition interactions with the receptor. Although the scoring of such interactions remains crude [34] , it is particularly helpful in such context to prioritize the chemistry effort on the promising fragment derivatives. The strength of this approach can also be its main limitation, when it is driven primarily by ease of synthetic chemistry and reagent accessibility, without maximizing opportunities to meet the structural constraints imposed by the receptor. Thus, the selected synthetic chemistry must take the targeted site into account even before docking of the resulting library, in particular regarding the linkers imposed by the chemistry. A more general approach would be to use the principles of de novo design methods [35,36] to guide elaboration of the fragment hits. This strategy is not restricted to a preselected chemistry or list of reagents, and computationally generates ideas directly selected to maximize their structural match to the receptor. Even under the receptor steric constraints, this approach faces a combinatorial explosion producing a vast number of suggestions. The associated chemical diversity and novelty may help, but the large array of generated ideas must be filtered aggressively on defined criteria. Pruning with calculated physico­ chemical descriptors relating to ADMET properties is straightforward and routine. However, reconciling the algorithmically generated ideas with tractable synthetic chemistry remains a major bottle neck for the de novo approach. Yet, starting from an experimentally discovered fragment core means that this key first step does not have to be tackled computationally. Thus, the de novo engine can operate directly on a validated fragment, to explore its substituents. So, www.future-science.com 1113 Commentary | Foloppe there is, in principle, a strong complementarity between FBDD and the de novo design strategy. Importantly, the de novo tools are very powerful idea generators. Even if not literally practical, such ideas can form the basis of fruitful discussions to inspire medicinal chemistry efforts. Growing versus linking Regardless of the precise techniques used to generate and prioritize ideas, there are two avenues to elaborate fragment hits into leads: growing and linking [37] . Growing refers to the stepwise addition of substituents to a fragment anchor, to target as efficiently as possible the surrounding binding site. Our experience, and discussions with other practitioners, indicate that growing is the most frequent situation, if only because linking requires that two fragments bind to adjacent sites on the target. This second situation is, however, sometimes made possible by the small size of the fragments, and the nature of the target [4,28,37,38] . Ideally, one should obtain an x-ray structure with both fragments bound simultaneously, instead of the fragments crystallized separately, as the binding of one fragment may influence the other [28] . When presented with two fragments bound in proximal pockets, the linking approach attempts to find tethers between them [39] . Examples of linking between fragments have been reported [4,28,37,38] . Theoretically, linking has long been recognized as a way to gain binding affinity over that of each individual constituent fragment, by reducing the penalty arising from the loss in rotational and translational entropy upon binding [28,40–42] . Linked fragments can indeed result in more potent compounds [28,37,38] , even without apparent favorable interactions between the linker and receptor [38] . Of course, the linker still needs to be sterically adequate, and selected such that it does not incur internal strain in its bioactive binding mode [28,37,38] . Such intramolecular conformational energies can be calculated rather accurately by modern computational chemistry [43] , but predicting the other contributions to the free Bibliography 1 2 Rees DC, Congreve M, Murray CW, Carr R. Fragment-based lead discovery. Nat. Rev. Drug Discov. 3, 660–672 (2004). Hubbard RE, Davis B, Chen I, Drysdale MJ. The SeeDs approach: integrating fragments into drug discovery. Curr. Top. Med. Chem. 7, 1568–1581 (2007). 1114 energy of binding remains challenging [42] . Yet, free-energy calculations were reported to have contributed to some fragment evolutions [11] . The strong complementarity that exists, in principle, between FBDD and perturbation-based rigorous free-energy calculations may become practical in the future. Future perspective Overall, FBDD has established itself as a powerful paradigm for the discovery of medicinal leads with desirable properties. With this strategy, the H2L process is arguably as close as one can currently be to rational drug design. It offers realistic opportunities to obtain leads quickly while controlling their properties. Of course, trial and error is still part of FBDD, but structure-based design tools do guide fragment optimization. There are still ample productivity gains to be realized by developing more reliable computational design protocols, integrated with synthetic chemistry. Another major benefit of FBDD will be to deepen and sharpen our fundamental understanding of molecular recognition. Indeed, fragment elaboration yields vast amount of systematic experimental data combining x-ray structures of protein–ligand complexes and associated binding affinities and kinetics. The mining of such data to dissect the underlying physical chemistry of protein–ligand recognition has only begun. So, documenting well-characterized case studies of fragment elaboration remains invaluable. One hopes that the lessons learned will go beyond empiricism and will ultimately inform the development of more powerful design tools. Financial & competing interests disclosure The author is an employee of Vernalis R&D Ltd, a pharmaceutical company that uses fragment-based drug design as its discovery platform. The author has no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed. No writing assistance was utilized in the production of this manuscript. 3 de Esch IJP. The HTS dogma is shattered, and now there are fragments everywhere. Drug Discov. Today Technol. 7, e147-e148 (2010). 4 Hajduk PJ, Greer J. A decade of fragmentbased drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 6, 211–219 (2007). Future Med. Chem. (2011) 3(9) 5 Fragment-Based Drug Design. Tools, Practical Approaches, and Examples. Kuo LC (Ed.). Elsevier, San Diego, CA, USA (2011). 6 Card GL, Blasdel L, England BP et al. A family of phosphodiesterase inhibitors discovered by cocrystallography and scaffold-based drug design. Nat. Biotechnol. 23, 201–207 (2005). future science group The benefits of constructing leads from fragments hits 7 8 9 10 11 Congreve M, Carr R, Murray C, Jhoti H. A ‘Rule of Three’ for fragment-based lead discovery? Drug Discov. Today 8, 876–877 (2003). 20 Edink E, Jansen C, Leurs R, de Esch IJP. Congreve M, Chessari G, Tisi D, Woodhead AJ. Recent developments in fragment-based drug discovery. J. Med. Chem. 51, 3661–3680 (2008). 21 Whittaker M, Law RJ, Ichihara O, Hesterkamp T, Hallett D. Fragments: past, present and future. Drug Discov. Today Technol. 7, e163-e171 (2010). 22 Hann MM, Leach AR, Harper G. Molecular Chessari G, Woodhead AJ. From fragment to clinical candidate - a historical perspective. Drug Discov. Today 14, 299–307 (2009). 23 Chen I, Hubbard RE. Lessons from Artis DR, Lina JJ, Zhanga C et al. Scaffoldbased discovery of indeglitazar, a PPAR pan-active anti-diabetic agent. Proc. Natl Acad. Sci. USA 106, 262–267 (2009). complexity and its impact on the probability of finding leads for drug discovery. J. Chem. Inf. Comput. Sci. 41, 856–864 (2001). fragment library design: ana­lysis of output from multiple screening campains. J. Comput. Aided Mol. Des. 23, 603–620 (2009). 24 Hopkins AL, Groom CR, Alex A. Ligand efficiency: a useful metric for lead selection. Drug Discov. Today 9, 430–431 (2004). 12 Hajduk PJ, Galloway WRJD, Spring D. A question of library design. Nature 470, 42–43 (2011). 25 Hajduk PJ. Fragment-based drug design: how big is too big? J. Med. Chem. 49, 6972–6976 (2006). 13 Brough PA, Barril X, Borgognoni J et al. Combining hit identification strategies: fragment-based and in Silico approaches to orally active 2-aminothieno[2,3-d]pyrimidine inhibitors of the Hsp90 molecular chaperone. J. Med. Chem. 52, 4794–4809 (2009). 14 15 Hubbard RE, Murray JB. Experiences in fragment-based lead discovery. In: FragmentBased Drug Design. Tools, Practical Approaches, and Examples, Kuo LC (Ed.). Elsevier Inc., Amsterdam 509–531 (2001). Murray CW, Blundell TL. Structural biology in fragment-based drug design. Curr. Opin. Struct. Biol. 20(497–507), (2010). 16 Michne WF. Hit-to-lead chemistry: a key element in new lead generation. Pharmaceutical News 3, 19–21 (1996). 17 Baurin N, Aboul-Ela F, Barril X et al. Design and characterization of libraries of molecular fragments for use in NMR screening against protein targets. J. Chem. Inf. Comput. Sci. 44, 2157–2166 (2004). 18 19 Retra K, Irth H, van Muijlwijk-Koezen JE. Surface plasmon resonance biosensor ana­lysis as a useful tool in FBDD. Drug Discov. Today Technol. 7, e181–e187 (2010). Antonysamy SS, Aubol B, Blaney J et al. Fragment-based discovery of hepatitis C virus NS5b RNA polymerase inhibitors. Bioorg. Med. Chem. Lett. 18, 2990–2995 (2008). future science group 33 Pierce AC, Rao G, Bemis GW. The heat is on: thermodynamic ana­lysis in fragment-based drug discovery. Drug Discov. Today Technol. 7, e189–e201 (2010). Blundell TL, Jhoti H, Abell C. Highthroughput crystallography for lead discovery in drug design. Nat. Rev. Drug Discov. 11, 45–54 (2002). 26 Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv. Drug Deliv. Reviews 23, 3–25 (1997). 27 Potter A, Oldfield V, Nunns C et al. Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution. Bioorg. Med. Chem. Lett. 20, 6483–6488 (2010). 28 Barker JJ, Barker O, Courtney SM et al. Discovery of a novel Hsp90 inhibitor by fragment linking. ChemMedChem 5, 1697–1700 (2010). 29 Barker JJ, Barker O, Boggio R et al. Fragment-based identification of Hsp90 inhibitors. ChemMedChem 4, 963–966 (2009). 30 Tsai J, Lee JT, Wang W et al. Discovery of a selective inhibitor of oncogenic B-Raf kinase with potent antimelanoma activity. Proc. Natl Acad. Sci. USA 105, 3041–3046 (2008). 31 Hajduk PJ. Puzzling through fragment-based drug design. Nat. Chem. Biol. 2, 658–659 (2006). 32 Babaoglu K, Shoichet BK. Deconstructing fragment-based inhibitor discovery. Nat. Chem. Biol. 2, 720–723 (2006). www.future-science.com | Commentary BREED: generating novel inhibitors through hybridization of known ligands. Application to CDK2, P38, and HIV protease. J. Med. Chem. 47, 2768–2775 (2004). 34 Ferrara P, Gohlke H, Price DJ, Klebe G, Brooks CLI. Assessing scoring functions for protein–ligand interactions. J. Med. Chem. 47, 3032–3047 (2004). 35 Schneider G, Fechner U. Computer-based de novo design of drug-like molecules. Nat. Rev. Drug Discov. 4, 649–663 (2005). 36 Loving K, Alberts I, Sherman W. Computational approaches for fragmentbased and de novo design. Curr. Top. Med. Chem. 10, 14–32 (2010). 37 Hung A, Silvestre H, Wen S, Ciulli A, Blundell T, Abell C. Application of fragment growing and fragment linking to the discovery of inhibitors of Mycobacterium tuberculosis pantothenate synthetase. Angew. Chem. Int. Ed. 48, 8452–8456 (2009). 38 Chung S, Parker JB, Bianchet M, Amzel M, Stivers JT. Impact of linker strain and flexibility in the design of a fragment-based inhibitor. Nat. Chem. Biol. 5, 407–413 (2009). 39 Thompson DC, Denny RA, Nilakantan R, Humblet C, Joseph-McCarthy D, Feyfant E. CONFIRM: connecting fragments found in receptor molecules. J. Comput. Aided Mol. Des. 22, 761–772 (2008). 40 Jencks WP. On the attribution and additivity of binding energies. Proc. Natl Acad. Sci. USA 78, 4046–4050 (1981). 41 Murray CW, Verdonk ML. The consequences of translational and rotational entropy lost by small molecules on binding to proteins. J. Comput. Aided Mol. Des. 16, 741–753 (2002). 42 Foloppe N, Hubbard R. Towards predictive ligand design with free-energy based computational methods? Curr. Med. Chem. 13, 3583–3608 (2006). 43 Foloppe N, Chen I. Conformational sampling and energetics of drug-like molecules. Curr. Med. Chem. 16, 3381–3413 (2009). 44 Roughley SD, Hubbard RE. How well can fragments explore accessed chemical space? A case study from heat shock protein 90. J. Med. Chem. 54, 3989–4005 (2011). 1115









