Multinomial Distribution Then I
advertisement
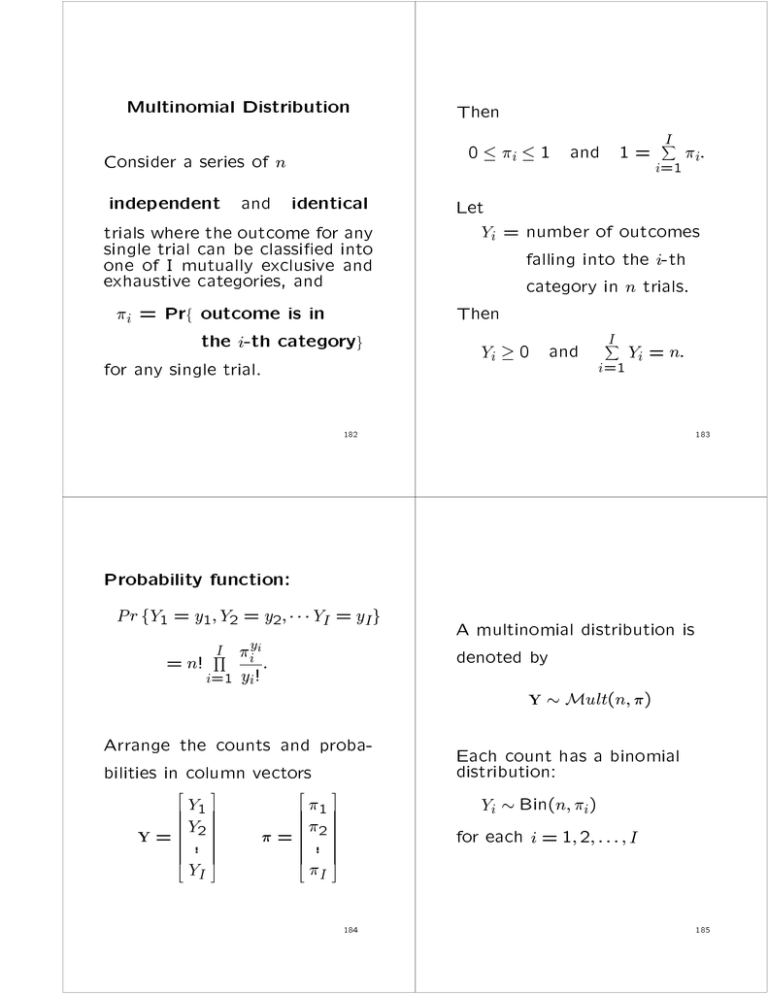
Multinomial Distribution
Consider a series of n
independent and identical
trials where
the beoutcome
for into
any
single
trial
can
classied
one of I mutually
exclusive
exhaustive
categories,
and and
i = Prf outcome is in
the i-th categoryg
for any single trial.
Then
I
0 i 1 and 1 = i=1
i:
Let
Yi = number of outcomes
falling into the i-th
category in n trials.
Then
I
Yi 0 and
Yi = n:
i=1
X
X
182
183
Probability function:
= y1; Y2 = y2; YI = yI g
I iyi
= n! i=1 yi! .
P r fY1
Y
Arrange the counts and probabilities in column vectors
2
Y
6
6
6
6
6
6
6
6
6
4
Y1
Y2
= .
YI
3
2
7
7
7
7
7
7
7
7
7
5
6
6
6
6
6
6
6
6
6
4
1
2
= .
I
3
7
7
7
7
7
7
7
7
7
5
184
A multinomial distribution is
denoted by
Y Mult(n; )
Each
count
has
a
binomial
distribution:
Yi Bin(n; i)
for each i = 1; 2; : : : ; I
185
Moments:
( ) = ni
V (Yi) = ni(1
Cov(Yi; Yj ) =
Observed (or sample)
proportions:
2
1
p= Y=
n
6
6
6
6
6
6
6
6
6
6
4
Y1=n
Y2=n
E Yi
3
2
7
7
7
7
7
7
7
7
7
7
5
6
6
6
6
6
6
6
6
6
4
p1
p2
. = .
YI=n
pI
3
7
7
7
7
7
7
7
7
7
5
i
)
nij
( ) = i
V (pi) = n1 i(1 i)
Cov(pi; pj ) = nij
E pi
187
186
where
Covariance matrix:
( )
V Y
2
1(1 1)
1 2
6
(1
2)
6
2 1
2
= n6
..
..
4
.
.
I 1
2
=n
6
6
6
6
6
4
1
2
I (1
..
.
I )
...
I
6
6
6
6
6
4
.
I 1
0
1I
2I
. ... .
3
7
7
7
5
= diag() =
Then
7
7
7
7
7
5
12 12
2
n 21 2
= n 1 I
2 I
3
2
...
I2
V
3
7
7
7
7
7
5
188
2
6
6
6
6
6
6
6
6
6
4
1
3
2
...
(p) = V (n1Y)
= n1 2 V (Y)
= n1 0
0
1
@
A
I
!
189
7
7
7
7
7
7
7
7
7
5
Limiting normal distribution:
For the j-th trial dene
0
0.
Yj =
1.
0
2
3
6
6
6
6
6
6
6
6
6
6
6
4
7
7
7
7
7
7
7
7
7
7
7
5
I 1
and
V
one in the i-th position and zeros elsewhere if the outcome
for the j-th trial is in
the i-th category
The Yj 's are i.i.d. random
vectors with
E (Yj ) = (Yj ) = The vector of sample
proportions
p1
p= .
= n1 n
2
3
6
6
6
6
6
4
7
7
7
7
7
5
pI
X
j
=1
0
Yj
= n1 Y
is a vector of sample means.
190
191
Likelihood function:
By the Multivariate Central
L
Limit Theorem
pn(p
) dist'n
! N (0; iYi
Yi
( ; Y) = n! i=1 !
I
0
as n ! 1
)
Y
Log-likelihood:
I
( ; Y) = log(n!) i=1
log(Yi!)
+ i=1I Yi log(i)
`
X
X
192
193
Method of Lagrange
multipliers:
Maximum likelihood
estimates (mle's):
Maximize
I
g (; ) = `(; Y)+ 1
i
i=1
Given observed counts
2
y1
y= .
6
6
6
6
6
4
yI
0
X
B
@
3
7
7
7
7
7
5
maximize `(; y) subject to the
I
constraint 1 = i=1
i .
X
= 1; 2; : : : ; I
Note that the mle's satisfy the
parameter constraints, i.e.,
I
1 = i=1
^i
.
X
196
:
X
195
Example:
i
C
A
Solve the likelihood equations
0 = @@gi = Yii i = 1; 2; : : : ; I
@g
0 = @
= 1 i=1I i
194
The mle's are
Y
^i = i = pi
n
1
Summer Squash
Sinnott and Durham (1922)
, 177{186.
Journal
of Heredity, 13
Results for n = 205 progeny of
cross bred white and yellow summer squash.
155 white
y1
y = y2 =
40 yellow
10 green
y3
2
3
2
3
6
6
6
6
6
4
7
7
7
7
7
5
6
6
6
6
6
4
7
7
7
7
7
5
197
The genetic model they considered suggests that
white: yellow: green
should occur with a ratio of
12 : 3 : 1:
Is this an appropriate model?
Test the null hypothesis
H0 : = 0
where
0;1
12=16
0 = 0;2 = 3=16
0;3
1=16
2
3
2
3
6
6
6
6
6
4
7
7
7
7
7
5
6
6
6
6
6
4
7
7
7
7
7
5
against the general alternative
HA : 0 < i < 1
3 = 1:
for i = 1; 2; 3 and i=1
i
X
198
1. Expected counts when H0 is
true
m
0=
=
m0;1
m0;2
m0;3
n0
2
3
6
6
6
6
6
4
7
7
7
7
7
5
12=16
= 205 3=16
1=16
153:78
= 38:4275
12:8125
2
3
6
6
6
6
6
4
7
7
7
7
7
5
2
3
6
6
6
6
6
4
7
7
7
7
7
5
200
199
2. Maximum likelihood estimates of \expected" counts
under HA.
m
^ A;1
m
^ A = m^ A;2
m
^ A;3
= np
155
= y = 40
10
2
3
6
6
6
6
6
4
7
7
7
7
7
5
2
3
6
6
6
6
6
4
7
7
7
7
7
5
201
3. Log-likelihood ratio test
(deviance)
3 y log m^ A;i
G2 = 2
i
m0;i
i=1
X
0
1
B
B
@
C
C
A
3 y log yi
= 2 i=1
i
m0;i
X
0
1
B
B
@
C
C
A
= 0:74
4. Pearson statistic
3 (^mA;i m0;i)2
X2 =
m0;i
i=1
2
= 3 (yi m0;i) = 0:69
X
X
=1
i
m0;i
Each statistic should be compared against the percentiles of
a central chi-squared distribution with
Dimension of
d.f. = parameter space
under HA
Dimension of
parameter space
under H0
=2 0
=2
2
3
6
6
6
6
6
4
7
7
7
7
7
5
2
3
6
6
6
6
6
4
7
7
7
7
7
5
202
SAS code
203
/* Establish a format to attach
labels to levels
of the color variable */
/* This program is stored in
the file
multfit.sas */
/* This program uses PROC IML in SAS
to test the fit of a completely
specified multinomial model against
a general alternative. It is applied
to test the fit of a genetic model
for color of summer squash (Sinnott
and Durham, 1922, Journal of Heredity,
13, 177-186). */
proc format;
value ccode 1 = 'White'
2 = 'Yellow'
3 = 'Green';
run;
/* Print the data set with
color labels */
proc print data=set1;
format color ccode.;
run;
data set1;
input color count;
cards;
1 155
2 40
3 10
run;
/* Use the IML procedure to compute
likelihood ratio and Pearson
chi-squared tests of the null
hypothesis that white:yellow:green
colors occur with a 12:3:1 ratio.*/
204
205
proc iml;
start multfit;
/* Enter the data */
use set1;
read all into w;
/* Smooth observed counts toward the
null hypothesis to avoid computing
the log of zero */
a = .000000001;
xl = (1-a)*x + a*m;
/* Create a column of counts */
x = w[ ,2];
/* Compute the deviance, df,
and a p-value */
g2 = 2*sum(x#log(xl/m));
df = nc-1;
pg2 = 1-probchi(g2,df);
/* Compute the total sample size */
n = sum(x);
/* Compute the number of categories */
nc = nrow(x);
/* Compute the Pearson statistic */
x2 = sum(((x-m)##2)/m);
px2 = 1-probchi(x2,df);
/* Enter the null hypothesis */
pi = {12, 3, 1};
pi = pi/sum(pi);
/* Compute expected counts */
m = n*pi;
207
206
/* Round off results and
print results */
g2 = round(g2,.001); pg2=round(pg2, .0001);
x2 = round(x2,.001); px2=round(px2, .0001);
print,,,, 'Tests of the null hypothesis; ', pi;
print,,, 'Observed Counts
Expected counts';
print x m;
print,,, '
Test'
'
DF
P-value';
print 'Deviance test: ' g2 df pg2;
print 'Pearson test: ' x2 df px2;
Obs
color
count
1
2
3
White
Yellow
Green
155
40
10
Tests of the null hypothesis;
PI
finish;
0.75
0.1875
0.0625
run multfit;
208
209
Observed Counts
X
M
155
40
10
153.75
38.4375
12.8125
Test
DF
/* Use the FREQ procedure in SAS to test
a null hypothesis for a multinomial
distribution. This code is posted as
multfit2.sas
*/
P-value
G2
DF
PG2
Deviance test: 0.741
2
0.6904
X2
DF
PX2
0.691
2
0.7078
Pearson test:
SAS code
Expected counts
data set1;
input type $9. y;
datalines;
white 155
yellow 40
green 10
run;
/* Note that the probabilities in the
null hypothesis are listed in the
order (green, white, yellow) because
SAS alphabetically orders the values
of the type variable. */
proc freq data=set1;
table type / testp = (.0625 .75 .1875);
weight y;
run;
211
210
S-PLUS code
The FREQ Procedure
type
Frequency
green
white
yellow
10
155
40
Percent
Test
Percent
4.88
75.61
19.51
6.25
75.00
18.75
Chi-Square Test
for Specified Proportions
Chi-Square
DF
Pr > ChiSq
#
#
#
#
#
#
#
#
This file contains Splus code for
testing the fit of a completely
specified multinomial model against
a general alternative. It is used
to test the fit of a genetic model
for the color of summer squash
(Sinnott and Durham, 1922, Journal
of Heredity, 13, 177-186).
#
The file is stored as
#
Enter the observed counts
multfit.ssc
x<-c(155, 40, 10)
0.6911
2
0.7078
#
Compute total sample size
n<-sum(x)
Sample Size = 205
#
Enter labels
labels<-c("white", "yellow", "green")
212
213
# Enter the hypothesized proportions
pi<-c(12, 3, 1)
pi<-pi/sum(pi)
# Display results
# Compute expected counts
e<-n*pi
# Smooth observed counts toward the null
# hypothesis to avoid computing the log
# of zero
a<-.000000001
xl<-(1-a)*x + a*e
g2<-round(g2, 3)
pg2<-round(pg2, 4)
x2<-round(x2, 3)
px2<-round(px2, 4)
dat<-as.matrix(cbind(labels, x, e))
nc<-ncol(dat)
dimnames(dat)[[2]]<c("labels", "observed", "expected")
cat(dimnames(dat)[[2]], format(t(dat)),
file="",
sep=c(rep(" ", nc-1), "\n"))
cat("\nDeviance test:", format(g2),
" df = ", format(df),
" p-value = ", format(pg2), "\n")
cat("\n Pearson test:", format(x2),
" df = ", format(df),
" p-value = ", format(px2), "\n")
# Compute the deviance
g2<-2*sum(x*log(xl/e))
df<-length(x)-1
pg2<-(1-pchisq(g2, df))
# Compute the Pearson statistic
x2<-sum(((x-e)**2)/e)
px2<-(1-pchisq(x2, df))
215
214
Displaying multinomial
counts in a two-way
contingency table
labels observed expected
white
155 153.75
yellow
40 38.4375
green
10 12.8125
Coronary Disease
and Serum
Cholesterol
An =simple
random
sample wasof
1329
patient
records
taken
from
records
of
a
specic
age/sex
group maintained by a
large HMO.
Example:
Deviance test:
0.741 df = 2 p-value = 0.6904
Pearson test:
0.691 df = 2 p-value = 0.7078
216
217
A 2 2 Contingency table: (n = 1329)
Each patient was classied into one of
four possible categories dened by two
traits (or factors).
Factor 1: Level of serum cholesterol
(i = 1) less than 220mg/100cc
(i = 2) at least 220mg/100cc
Factor 2: Coronary disease status
(j = 1) Present
(j = 2) Absent
Coronary Disease
Present
<
Absent
220
y11 = 20 y12 = 553
220
y21 = 72 y22 = 684
Serum
Cholesterol
(mg/100g)
Rearrange the counts into a column
vector
2
Y
=
6
6
6
6
6
4
218
Y11
Y12
Y21
Y22
3
7
7
7
7
7
5
Mult(n; )
219
Question:
where
2
=
and
1=i
2
X
2
X
j
=1 =1
ij
6
6
6
6
6
4
11
12
21
22
and
3
7
7
7
7
7
5
n
=
XX
i j
Yij
Here
= proportion of HMO patients
with serum cholesterol less
than 220mg/100cc and no
coronary disease
12
220
Is the incidence of coronary
disease the same for both
cholesterol categories?
Test the t of the
independence model. Here
\independence" means
that the incidence of
coronary disease is the
same for each serum
cholesterol category.
221
With respect to the elements of
2
The null hypothesis can be expressed in terms of conditional
probabilities:
H0
8
<
: Pr
:
8
<
= Pr
:
coronary low cholesterol
disease
level
9
=
;
coronary high cholesterol
disease
level
9
=
;
=
6
6
6
6
6
6
6
6
6
4
11
12
21
22
3
7
7
7
7
7
7
7
7
7
5
this is written as
11
21
H0 :
=
11 + 12 21 + 22
An equivalent statement is
H0 : ij = i++j
where
i+ = j ij and +j = i ij
P
P
223
222
The vector of proportions is a
function of the parameters
1+
2+
+1
+2
Likelihood
Y
2
2
ijij
L(; Y) = n!
i=1 j =1 Yij !
Y
Note that
1 = 1+ + 2+
and
1 = +1 + +2
Then, is a function of just two
parameters,
f1+; +1g:
224
Y
Find maximum likelihood
estimates by maximizing
g (11; 12; 21; 22; )
= log(n!) i j log(Yij !)
+ i j Yij log(ij )
+(1 i j ij )
X X
X X
X X
225
Solve the equations
0 = @@gij = Yijij i
j
= 1; 2
= 1; 2
0 = @@g = 1 i j ij
Solution
Y
^ij = ij
i = 1; 2 j = 1; 2
n
= n
mle's for expected counts
n^ij = Yij
X X
Maximum likelihood estimates
for expected counts under
independence
Substitute ij = i j into the
likelihood function to obtain
n! i
j
Y
+
Q
Yij
ij
Q
2
2
=1
=1
= n!
=
Q
Q
+
ij !
Y
( i+ +j ) ij
Q
2
2
=1
=1
ij !
i
2
i=1
j
n!
Q
2
j =1
Y
Y
Y
Q
2
i+ Q2 +j
Yij i=1 i+ j =1 +j
227
226
Maximize
g (1+; +1; 2+; +2; 1; 2)
= log(n!) i j log(Yij !)
+ i Yi+ log(i+)
+ j Y+j log(+j )
+1(1 i i+)
+2(1 j +j )
X X
X
X
X
X
228
Solve the equations
0 = @@gi+ = Yii++ 1 i = 1; 2
0 = @@g+j = Y++jj 2 j = 1; 2
0 = @@g1 = 1
0 = @@g2 = 1
X
i
X
j
i+
+j
229
Solution:
Y
^i+ = i+
n
Y
^+j = +j
n
= 1; 2
j = 1; 2
i
The \expected counts" are
Then the m.l.e.'s for the cell
proportions and expected counts
are
Y+j
Y
^ij = ^i+^+j = i+
n
n
0
1
0
1
@
A
@
A
Serum
chol.
Coronary Disease
Present
Absent
< 220
(573)(92)
= 39:67
m
^ 11 =
1329
(573)(1237)
= 533:33
m
^ 12 =
1329
y1+ = 573
< 220
m
^ 21 =
(756)(1237)
= 703:62
1329
y2+ = 756
(756)(92)
= 52:33
1329
m
^ 22 =
y+1 = 92
y+2 = 1237
^ = n ^ij = Yi+nY+j
mij
230
The \t" of the independence
model is assessed by comparing
it to the \general alternative"
model that places no restrictions
on other than
1 = i j ij :
X X
The m.l.e.'s for the expected
counts are the observed counts,
m
^ A;ij = Yij :
232
231
Compute
G2 = 2
Yij log Yij =m
^ ij = 19:8
i j
2 ^ ij = 18:4
X2 =
(
Yij m
^
ij ) =m
i j
with d.f. = 3 2 = 1:
Since 2(1):005 = 7:88, it appears
that
the independence
model ofis
inappropriate.
The
incidence
coronary
disease
is higher group.
for the
higher serum
cholesterol
X X
X X
233
Comparing vectors of proportions for several independent
samples (or experiments):
Suppose j = 1; 2; : : : ; J
simple random samples (or
experiments) are done. For the
j -th survey (or experiment)
the nj outcomes are classied
into I categories. The random
counts are
2
Yj
6
6
6
6
6
6
6
6
6
4
Y1j
Y2j
= .
YIj
3
7
7
7
7
7
7
7
7
7
5
Mult(nj ; j )
where
2
j
and
6
6
6
6
6
6
6
6
6
4
= .
Y
1; Y2; ; YJ
are independent vectors of random counts.
Also dene
2
Pj
6
6
6
6
6
6
6
6
6
4
P1j
P2j
3
2
7
7
7
7
7
7
7
7
7
5
6
6
6
6
6
6
6
6
6
6
4
= . =
PIj
Y1j=nj
Y2j=nj
.
YIj=nj
3
7
7
7
7
7
7
7
7
7
7
5
= n1j Yj :
236
Ij
3
7
7
7
7
7
7
7
7
7
5
1 = i=1I ij
X
and
nj
= i=1I Yij
X
for j = 1; :::; J:
234
No outcome of any experiment
has any inuence on any other
outcome;
1j
2j
235
The hypothesis
H0 : 1 = 2 = = J
is often of interest. This is often
called the homogeneity or
independence model and it is
usually compared to the general
alternative that only assumes
I
ij = 1
i=1
for each j = 1; ; J .
X
237
Example:
The General Social Survey, conducted
by the National Opinion Research at
the University of Chicago, uses many of
the same questions from year to year.
Haberman (1978) examined responses
to the question
\In general, do you think
courts in this area deal
too harshly or not harshly
enough with criminals?"
Observed Counts
Year of Survey
Response
1972
1973
1974
1975
Too harshly (i=1)
105
68
42
61
About right (i=2)
265
196
72
144
1066
1092
580
1174
173
138
51
104
4
10
8
7
1612
1504
753
1490
Not harshly
enough (i=3)
Don't know (i=4)
No answer (i=5)
Sample Size
for
independent
1973,
1974, andsurveys
1975. taken in 1972,
238
Maximum likelihood estimation for expected counts:
Percentages
Year of Survey
Response
1972
1973
1974
1975
Too harshly (i=1)
6.5
4.5
5.6
4.1
About right (i=2)
16.4
13.0
9.6
9.7
enough (i=3)
66.1
72.6
77.0
78.8
Don't know (i=4)
10.7
9.2
6.8
7.0
0.3
0.7
1.1
0.5
1612
1504
753
1490
Not harshly
No answer (i=5)
239
Model A: general alternative
Yj Mult(nj ; j ) j = 1; 2; 3; 4
are independent vectors of random counts and
5 =1
1T j =
ij
i=1
for each j = 1; 2; 3; 4 .
X
Sample Size
240
241
log-likelihood function:
The joint likelihood function is
L(1; 2; 3; 4; Y1; Y2; Y3; Y4)
4 n! 5
= j=1
j
!
i=1
This is sometimes
\product
multinomial"called
model.the
2
Y
6
6
6
6
4
Y
Y
ijij
Yij
3
7
7
7
7
5
4 log(n !) 5 4 log(Y !)
j
ij
j =1
i=1 j =1
4 5 Y log( )
+ j=1
ij
ij
i=1
X
X
X
X
X
Maximize this with respect to
the conditions
5 = 1 for
ij
i=1
j
X
242
Dene
(
g 1; 2; 3; 4; 1; 2; 3; 4
4 log(n !)
= j=1
j
)
0 = @@gij
= Yijij
4 5 log(Y !)
ij
j =1 i=1
4 5 Y log( )
+ j=1
ij
ij
i=1
4 1 5 .
+ j=1
j
ij
i=1
X
X
X
B
@
5 = 1 i=1
ij
X
1
X
j
i
j
= 1; 2; : : : ; 5
= 1; : : : ; 4
0 = @@gj
X
0
243
Solve
X
X
= 1; 2; 3; 4
C
A
244
j
= 1; : : : ; 4
245
Model B: Independence (or
homogeneity)
model
Solution:
or
^ = pij = Yij =nj
ij
2
^j = pj =
1
Y
nj j
expected count = nj ^ij
= Yij
= observed count:
Substitute for j in the
log-likelihood subject to
5 = 1T :
1 = i=1
i
X
247
246
Dene
4 log(n !)
g (; ) =
j
j =1
X
4 5 log(Y !)
ij
j =1 i=1
5 Y log( )
+ i=1
i
i+
5 + 1 i=1
i
X
X
X
0
B
@
1
X
1
H0 : 1 = 2 = 3 = 4 = = .
6
6
6
6
6
4
C
A
248
Solve
0 = @@gi = Yi+i for i = 1; 2; : : : ; 5
and
5 @g
0 = @
= 1 i=1
i
X
249
5
3
7
7
7
7
7
5
Test
H0 : 1 = 2 = 3 = 4 (model B)
against the general alternative
(model A).
Solution:
4 n
^i = Yi+/
j
j =1
X
or
4 np 4 n :
^ = j=1
j j/
j
j =1
X
X
Compute:
2
(
yij m
^
)
ij
2
X =
m
^ ij = 87:4
i j
Expected counts are:
m
^ ij = nj ^i = n4j Yi+
X
j
=1
X X
nj
column row
= totaltotal fortotal
entire table
2
3
2
3
6
4
7
5
6
4
7
5
2
3
6
4
7
5
G2
=2i
0
X X
j
yij
1
log ^ = 87:1
B
B
@
yij
mij
C
C
A
each with 16 4 = 12 d.f.
250
Conclusion:
The proportion of the population in at least one category was
not the same in all 4 years:
In which categories
or years
did changes occur?
Examine
patterns in observed
proportions
dierences between observed
and expected counts
252
251
Pearson residuals
X
m
^ ij
rij = ij
m
^ ij
r
Note that X 2 = i j rij2
X X
adjusted residuals
Xij m
^ ij
=
rij
m
^ ij [1 ^ij ][1
r
]
nj =n
J
where n = j=1
nj
X
253
Adjusted Residuals
Observed Proportions
Overall
Response
Too harshly
About right
1972
1973
1974
1975
Proportion
6.5
4.5
5.6
4.1
5.2
9.6
9.7
12.6
16.4
13.0
Not harshly
enough
66.1
72.6
10.7
Don't know
No answer
77.0
9.2
78.8
6.8
73.0
7.0
8.7
0.3
0.7
1.1
0.5
0.5
16.13
1504
753
1490
5360
From 1972 through 1974 there was an
increase in the proportion of the population that felt the courts do not deal
harshly enough with criminals and a
corresponding decrease in the proportion of \about right" and \don't know"
opinions.
Response
1972
1973
1974
1975
Too harshly
3.0
-1.3
0.6
-2.2
About right
5.5
0.6
-2.7
-4.1
-7.5
0.4
2.7
5.9
3.5
0.8
-2.0
-2.8
-1.9
0.8
2.1
-0.4
Not harshly
enough
Don't know
No answer
Look at the
sign and size
of the adjusted residuals.
255
254
Compare 1974 to 1975
Observed
Counts
Response
Expected
Counts
1974
1975
1974
1975
Too harshly
42
61
34.58
68.42
About right
72
144
72.51
143.49
580
1174
588.84
1165.16
51
104
52.04
102.96
8
7
5.04
9.96
753
1490
753
1490
Not harshly
enough
Don't know
No Answer
Sample Size
X2
5 2 (yij m^ ij )2 = 5:3
= i=1
m
^ ij
j =1
X
X
Conclusion:
Essentially no changes
in the proportions of the
population holding various opinions between
1974 and 1975.
with 4 d.f. and p-value = :26.
256
257
SAS code
/* This program is stored in
the file crim1.sas */
Values of X 2 for testing
homogeneity for pairs of
years
/* Enter the data as counts
in a 2-dimensional table */
Second Year
First Year
1973
1972
21.3
1974
<.001)
(
1973
41.7
<.001)
(
data set1;
input response year count;
cards;
1 1 105
1 2 68
1 3 42
1 4 61
2 1 265
2 2 196
. . .
. . .
5 4
7
run;
1975
65.9
(
<.001)
12.0
16.5
(.016)
(.002)
1974
5.3
(2.62)
258
/* Use PROC FORMAT to assign labels
to the row and column categories */
proc format;
value rowfmt 1
2
3
4
5
value colfmt 1
2
3
4
=
=
=
=
=
=
=
=
=
259
/* Compare responses for each
pair of years without printing
tables of counts and proportions */
'Too harshly'
'About right'
'Too lenient'
'No opinion'
'No answer';
'1972'
'1973'
'1974'
'1975';
data set2; set set1;
if(year=1 or year=2);
title 'Comparison Between 1972 and 1973';
proc freq data=set2;
table response*year / chisq noprint;
weight count;
run;
/* Compute test for independence */
title 'Annual Opinions on Treatment of Criminals';
proc freq data=set1;
table response*year /
chisq expected cellchi2;
weight count;
format response rowfmt.
year colfmt.;
run;
260
data set2; set set1;
if(year=1 or year=3);
title 'Comparison Between 1972 and 1974';
proc freq data=set2;
table response*year / chisq noprint;
weight count;
run;
261
data set2; set set1;
if(year=3 or year=4);
title 'Comparison Between 1974 and 1975';
proc freq data=set2;
table response*year / chisq noprint;
weight count;
run;
data set2; set set1;
if(year=1 or year=4);
title 'Comparison Between 1972 and 1975';
proc freq data=set2;
table response*year / chisq noprint;
weight count;
run;
data set2; set set1;
if(year=2 or year=3);
title 'Comparison Between 1973 and 1974';
proc freq data=set2;
table response*year / chisq noprint;
weight count;
run;
data set2; set set1;
if(year=2 or year=4);
title 'Comparison Between 1973 and 1975';
proc freq data=set2;
table response*year / chisq noprint;
weight count;
run;
262
263
Comparison between 1973 and 1974
Statistic
Annual Opinions on Treatment of Criminals
The FREQ Procedure
Statistics for Table of response by year
DF
Chi-Square
Likelihood Ratio Chi-Square
Mantel-Haenszel Chi-Square
Phi Coefficient
Contingency Coefficient
Cramer's V
Value
Prob
4 21.2788 0.0003
4 21.4459 0.0003
1 7.4150 0.0065
0.0826
0.0823
0.0826
Sample Size = 3117
Statistic
DF
Value
Chi-Square
12 87.3596
Likelihood Ratio Chi-Square 12 87.0513
Mantel-Haenszel Chi-Square
1 11.1240
Phi Coefficient
0.1277
Contingency Coefficient
0.1266
Cramer's V
0.0737
Sample Size = 5360
Prob
<.0001
<.0001
0.0009
Comparison Between 1972 and 1974
Statistic
DF
Chi-Square
Likelihood Ratio Chi-Square
Mantel-Haenszel Chi-Square
Phi Coefficient
Contingency Coefficient
Cramer's V
Value
4 41.7255 <.0001
4 42.7808 <.0001
1 4.3917 0.0361
0.1328
0.1316
0.1328
Sample Size = 2366
264
Prob
265
Comparison Between 1972 and 1975
Statistic
DF
Chi-Square
Likelihood Ratio Chi-Square
Mantel-Haenszel Chi-Square
Phi Coefficient
Contingency Coefficient
Cramer's V
Value
Comparison Between 1973 and 1975
Prob
4 65.9007 <.0001
4 66.6726 <.0001
1 12.4171 0.0004
0.1457
0.1442
0.1457
Statistic
Chi-Square
Likelihood Ratio Chi-Square
Mantel-Haenszel Chi-Square
Phi Coefficient
Contingency Coefficient
Cramer's V
Sample Size = 3103
DF
Chi-Square
Likelihood Ratio Chi-Square
Mantel-Haenszel Chi-Square
Phi Coefficient
Contingency Coefficient
Cramer's V
Value
Prob
Statistic
4 16.5413 0.0024
4 16.5917 0.0023
1 0.5301 0.4665
0.0743
0.0741
0.0743
DF
Chi-Square
Likelihood Ratio Chi-Square
Mantel-Haenszel Chi-Square
Phi Coefficient
Contingency Coefficient
Cramer's V
Sample Size = 2257
4
4
1
S-PLUS code
#
#
#
#
#
Enter the data as a data frame.
This includes column headings
that identify the years and row
labels that identify the
type of response.
Compute Pearson chi-squared
ests for each pair of years
chisq.test(crim1.mat[ ,c(1,2)])
chisq.test(crim1.mat[ ,c(1,3)])
chisq.test(crim1.mat[ ,c(1,4)])
chisq.test(crim1.mat[ ,c(2,3)])
Store the data as a matrix object
chisq.test(crim1.mat[ ,c(2,4)])
chisq.test(crim1.mat[ ,c(3,4)])
crim1.mat<-as.matrix(crim1.dat)
#
#
#
#
#
#
#
#
#
List the table
crim1.mat
#
#
Compute the Pearson chi-squared
test for independence
chisq.test(crim1.mat)
268
Prob
5.2610 0.2615
5.0254 0.2847
0.4879 0.4848
0.0484
0.0484
0.0484
267
#
#
crim1.dat<-read.table("crim1.dat",header=T)
#
Value
Sample Size = 2243
266
This code is stored in the
file crim1.ssc
Prob
Comparison Between 1974 and 1975
4 12.0136 0.0173
4 12.2566 0.0155
1 0.0076 0.9307
0.0730
0.0728
0.0730
#
#
Value
Sample Size = 2994
Comparison Between 1973 and 1974
Statistic
DF
There does not seem to be a
corresponding built in function
to compute just the deviance.
The deviance can be obtained
from the glm function as follows.
We will first enter the data in
another way to form a two-way
contingency table with factor labels.
269
#
#
#
#
#
Enter the counts for the contingency
table. The function expand.grid( )
creates a dataframe containing all
combinations of its arguments: vectors,
factors, or lists.
crim1 <- cbind(expand.grid(
Year=c("1972","1973","1974","1975"),
Opinion=c("Too harshly","About right",
"Too lenient","No opinion","No answer")),
Fr=c(105, 68, 42, 61,
265, 196, 72, 144,
1066, 1092, 580, 1174,
173, 138, 51, 104,
4, 10, 8, 7))
#
#
#
#
Fit the independence model using the
glm( ) function. Note that a
Poisson distribution must be specified,
even though we have multinomial data.
crim1.indep <- glm(Fr ~ Opinion + Year,
family=poisson, data=crim1)
# Display the results
summary(crim1.indep, correlation=F)
# If you just want to see the
# value of the deviance use
deviance(crim1.indep)
#
#
#
#
The summary( ) function does not supply
p-values in this application. Use the
anova( ) function with test="Chisq" to
get p-values for the deviance tests.
anova(crim1.indep,test="Chisq")
# Use fitted( ) to compute the expected
# counts and compute the value of the
# Pearson chi-squared statistic
crim.fit <- as.matrix(fitted(crim1.indep))
crim.x <- as.matrix(crim1$Fr)
pearson <- apply((((crim.x - crim.fit)^2)/
crim.fit), 2, sum)
p <- 1.0 -pchisq(pearson,12)
270
271
This is the output from the Splus
code stored on the file crim1.ssc
> crim1.dat<-read.table("crim1.dat",header=T)
> crim1.mat<-as.matrix(crim1.dat)
> crim1.mat
# Display the results
cat("\n", "Value of the Pearson statistic: ",
pearson, "\n")
cat("\n", "P-value for the Pearson statistic: ",
p, "\n")
272
1972 1973 1974 1975
Too_harshly 105
68
About_right 265 196
Too_lenient 1066 1092
No_opinion 173 138
No_answer
4
10
42
61
72 144
580 1174
51 104
8
7
273
>chisq.test(crim1.mat[
>chisq.test(crim1.mat[
>chisq.test(crim1.mat[
>chisq.test(crim1.mat[
>chisq.test(crim1.mat[
>chisq.test(crim1.mat[
,c(1,2)])
,c(1,3)])
,c(1,4)])
,c(2,3)])
,c(2,4)])
,c(3,4)])
> chisq.test(crim1.mat)
Pearson's chi-square test without
Yates' continuity correction
Pearson's chi-square test without
Yates' continuity correction
data: crim1.mat
X-squared = 87.3596, df = 12, p-value = 0
Warning messages:
Expected counts < 5. Chi-squared
approximation may not be appropriate. in:
chisq.test(crim1.mat)
data: crim1.mat[, c(1, 2)]
X-squared = 21.2788, df = 4, p-value = 3e-04
Pearson's chi-square test without
Yates' continuity correction
data: crim1.mat[, c(1, 3)]
X-squared = 41.7255, df = 4, p-value = 0
Warning messages:
Expected counts < 5. Chi-squared
approximation may not be appropriate. in:
chisq.test(crim1.mat[, c(1, 3)])
274
Pearson's chi-square test without
Yates' continuity correction
data: crim1.mat[, c(1, 4)]
X-squared = 65.9007, df = 4, p-value = 0
Pearson's chi-square test without
Yates' continuity correction
data: crim1.mat[, c(2, 3)]
X-squared = 12.0136, df = 4, p-value = 0.0173
Pearson's chi-square test without
Yates' continuity correction
data: crim1.mat[, c(2, 4)]
X-squared = 16.5413, df = 4, p-value = 0.0024
Pearson's chi-square test without
Yates' continuity correction
data: crim1.mat[, c(3, 4)]
X-squared = 5.261, df = 4, p-value = 0.2615
276
275
> crim1 <- cbind(expand.grid(
Year=c("1972","1973","1974","1975"),
Opinion=c("Too harshly","About right",
"Too lenient",
"No opinion","No answer")),
Fr=c(105, 68, 42, 61,
265, 196, 72, 144,
1066, 1092, 580, 1174,
173, 138, 51, 104,
4, 10, 8, 7))
> crim1.indep <- glm(Fr ~ Opinion + Year,
family=poisson, data=crim1)
> summary(crim1.indep, correlation=F)
Call: glm(formula = Fr ~ Opinion + Year,
family = poisson, data = crim1)
Deviance Residuals:
Min
1Q
Median
3Q
Max
-3.361964 -1.86109 0.1317903 1.393514 4.100514
277
Coefficients:
(Intercept)
Opinion1
Opinion2
Opinion3
Opinion4
Year1
Year2
Year3
Value
4.55563937
0.44863522
0.73425598
-0.16477661
-0.65423965
-0.03498379
-0.24226735
0.04948287
> anova(crim1.indep,test="Chisq")
Std. Error
0.041097475
0.035704247
0.013040165
0.013087555
0.037270056
0.017921519
0.013536157
0.007751406
t value
110.849617
12.565318
56.307260
-12.590328
-17.554029
-1.952055
-17.897794
6.383728
(Dispersion Parameter for Poisson family
taken to be 1 )
Analysis of Deviance Table
Poisson model
Response: Fr
Terms added sequentially (first to last)
Df Deviance Resid. Df Resid. Dev Pr(Chi)
NULL
19 8251.947
Opinion 4 7771.211
15
480.735
0
Year 3 393.684
12
87.051
0
Null Deviance: 8251.947 on 19 degrees of freedom
Residual Deviance: 87.05134 on 12 degrees
of freedom
Number of Fisher Scoring Iterations: 3
> crim.fit <- as.matrix(fitted(crim1.indep))
>crim.x <- as.matrix(crim1$Fr)
>pearson <- apply((((crim.x - crim.fit)^2)
/crim.fit), 2, sum)
>p <- 1.0 -pchisq(pearson,12)
278
279
S-PLUS for Windows
>cat("\n", "Value of the Pearson statistic: ",
pearson, "\n")
>cat("\n", "P-value for the Pearson statistic: ",
p, "\n")
Value of the Pearson statistic: 87.359438070264
P-value for the Pearson statistic: 1.598721e-13
Click on Data ) Data select
) click on New Data and
enter a data le name in
the New Data box
) enter the data in the
spreadsheet that appears
with columns labeled
response year y
for response category, year
and count, respectively
Click on Statistics
) Data summmaries
) Crosstabulations
) make selections in the boxes
280
281
*** Crosstabulations ***
Call:
crosstabs(formula = y ~ response + year,
data = crim1, na.action = na.fail,
drop.unused.levels = T)
5360 cases in table
+----------+
|N
|
|N/RowTotal|
|N/ColTotal|
|N/Total |
+----------+
282
response|year
|1
|2
|3
|4
|RowTotl|
--------+--------+--------+--------+--------+-------+
1
| 105
| 68
| 42
| 61
|276
|
|0.3804 |0.2464 |0.1522 |0.221
|0.05149|
|0.0651 |0.04521 |0.05578 |0.04094 |
|
|0.01959 |0.01269 |0.007836|0.01138 |
|
--------+--------+--------+--------+--------+-------+
2
| 265
| 196
| 72
| 144
|677
|
|0.3914 |0.2895 |0.1064 |0.2127 |0.1263 |
|0.1643 |0.1303 |0.09562 |0.09664 |
|
|0.04944 |0.03657 |0.01343 |0.02687 |
|
--------+--------+--------+--------+--------+-------+
3
|1066
|1092
| 580
|1174
|3912
|
|0.2725 |0.2791 |0.1483 |0.3001 |0.7299 |
|0.6609 |0.7261 |0.7703 |0.7879 |
|
|0.1989 |0.2037 |0.1082 |0.219
|
|
--------+--------+--------+--------+--------+-------+
4
| 173
| 138
| 51
| 104
|466
|
|0.3712 |0.2961 |0.1094 |0.2232 |0.08694|
|0.1073 |0.09176 |0.06773 |0.0698 |
|
|0.03228 |0.02575 |0.009515|0.0194 |
|
--------+--------+--------+--------+--------+-------+
5
|
4
| 10
|
8
|
7
|29
|
|0.1379 |0.3448 |0.2759 |0.2414 |0.00541|
|0.00248 |0.006649|0.01062 |0.004698|
|
|7.463e-4|0.001866|0.001493|0.001306|
|
--------+--------+--------+--------+--------+-------+
ColTotal|1613
|1504
|753
|1490
|5360
|
|0.3009 |0.2806 |0.1405 |0.278
|
|
--------+--------+--------+--------+--------+-------+
Test for independence of all factors
Chi^2 = 87.35959 d.f.= 12 (p=1.598721e-013)
Yates' correction not used
Some expected values are less than 5, don't trust p-value
283


