S. cerevisiae growth on xylose L. Mande ; W.H. Van Zyl
advertisement

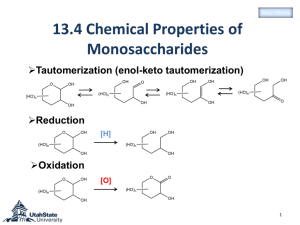
Evaluation of S. cerevisiae promoters during growth on xylose L. Mande1; W.H. Van Zyl2; I. Ncube1; D.C. La Grange1 Motivation •Identify a promoter that is strongly induced during growth on xylose • Increase production of recombinant enzymes needed during the degradation of lignocelluloses •Recombinant enzymes such as xylanases, beta- glucosidase, cellulases etc Introduction S.cerevisiae has been used for years in the production of recombinant proteins S.cerevisiae is a widely used organism - fast cell growth - tolerate high ethanol concentration - tolerate wide spectrum of inhibitors - well-characterised physiology & genetics - GRAS status Introduction Glucose is the most abundant sugar in nature (cellulose) S. cerevisiae is a Crabtree positive yeast Its produces ethanol on glucose under aerobic conditions (little biomass) High expression level of recombinant protein is linked to the amount of biomass obtained during fermentation (Ferndahl et al., 2010) Introduction Xylose is the second most abundant sugar in nature and considered a waste in most industries S. cerevisiae cannot utilise xylose as a carbon source S.cerevisiae engineered to grow on xylose has been reported to produce more biomass on xylose Xylose is the more suitable carbon source in recombinant protein production Introduction Promoter - ENO1, ENO2, YG100, PGK1, ADH2,GPD3 Reporter gene - Xylanase gene Terminator - ENO1, ENO2, YG100, PGK1, ADH2,GPD3 Episomal selectable marker - URA3 promoter Gene of interest Terminator Promoter Reporter gene Terminator Marker Engineering S.cerevisiae(Y294) sXI gene (B.thetaiotaomicron) XYL3 (P. stipitis) Fungi Bacteria D-xylose D-xylose NAD(P)H Xylose reductase NAD(P)+ Xylose isomerase D-xylitol GRE3 NAD+ Xylitol dehydrogenase NADH D-xylulose ATP Xylulokinase D-xylulose-5-P D-xylulose Xylulokinase ATP D-xylulose-5-P Restriction map of the plasmids SalI (7668) HindIII (7650) BamHI (7680) XhoI (7379) BglII (7372) XhoI (7229) PGK1t 1 EcoRI (6592) XYN2 PGK1p pP GK1 7724bp HindIII (5111) URA3 ApaI (4393) Figure 1. pPGK1 used as base for the construction of all episomal plasmids. Results and Discussion A B Figure 2. Engineered S.cerevisiae on xylose and SC –URA3 with glucose. Results and Discussion Growth of engineered and wild type S.cerevisiae on xylose Growth of engineered and wild type S.cerevisiae on glucose 16 20 18 14 16 12 12 OD600nm OD600nm 14 10 8 10 8 6 6 4 4 2 2 0 0 10 20 30 40 50 Incubation Time 60 70 80 0 0 50 100 150 200 Incubation Time (Hour) Figure 3.Growth curve of the wild type and engineered yeast on glucose and on xylose Results and Discussion Transformants spotted on RBB-xylan plates with different carbon source RBB-xylan xylose RBB-xylan glucose Figure 4. The RBB-xylan plates used to confirm xylanase activity Results and Discussion DNS ASSAY was done to determine the xylanase activity following the method by (Bailey et al, 1992) Figure 5. Determination of the amount of xylanase enzyme produced using DNS assay Results and Discussion Enzyme activity 250 Control 200 PGK1 nkat/ml 150 ENO1 100 ENO2 50 ADH2 0 0 50 100 150 200 Time (h) 250 300 Figure 6.. .Determination of the amount of xylanase enzyme produced using DNS assay Construction of fur1::LEU2 strains Glutamine De novo synthesis Orotidine 5'-P FUR1 Uridine 5'-P Uracil URA3 Uridine 5'-P Cytidine 5'-PPP DNA Kern et al., 1990 Construction of fur1::LEU2 strains pUC plasmid FUR1 1 FUR1 gene disruption 3.27 pDF1 fur1' LEU2 FUR1 gene replacement Yeast chr 8R FUR1 'fur1 1.33 2 3 FUTURE WORK Evaluation of episomal promoters during growth on xylose Determine metabolic burden during growth on xylose in the bioreactor ACKNOWLEDGEMENT Many thanks Dr. La Grange Prof Ncube and Prof van Zyl Dr V. Mbazima Van Zyl lab (Stellenbosch University) Department of BMBT (University of Limpopo) RSES for financial assistance Thank you ………………… Fur1 Disruption fur1::LEU2 allele was Isolated as 3.27kb from pDF1 plasmid NcoI-NsiI restriction enzymes Disrupted using the gene replacement method Introduction (Aristidou and Pentila,2000) Transformation of integrated plasmids Transformation - Transformation in S. cerevisiae S228c strain - Transformants selected on geneticin (G418) NotI 1 40-ura PGK1p Amp uPGK1x XhoI (1583) PGK1t NotI HindIII (3515) 40-ura G418 HindIII (2939) Promoter XYN2 PGK1 Marker PacI (1044) XYN2 5754bp SalI (3497) NotI (3490) U EcoRI (396) NotI (424) ApaI (473) U AscI (1723) EcoRI (2007) Integrating expression cassettes PGK1p and PGK1t isolated from plasmid by restriction digest G418 isolated from plasmid by PCR 40bp URA3 overhangs added to expression cassette (target cassette to URA3-locus) Expression cassette cloned into pUC19 U Promoter XYN2 PGK1 Marker U Transformation Rapid genomic DNA isolation, “Bust and Grab” method Construction of the strain that grows on xylose S. cerevisiae Y294 (Matα leu 2-3, 112 ura 3-52,his 3,trp1289) Transformation with pMJM121 with Synthetic codon optimised xylose isomerase B. thetaiotaomicron XI Selective marker (zeocin) Disruption cassette (gre3::Xyl3Hygromycin) was used to knock out GRE3 gene RESULTS U Promoter XYN2 Marker U Promoter XYN2 A PGK1t XYN2 B 10000 8000 6000 5000 ADH2 1735bp 4000 3500 3000 2500 2000 PFK2 1462bp 1500 XYN2 610 bp 1000 750 PFK1 909bp GAL10 1228bp ENO1 897bp 500 250 Figure 2: (A) Agarose gel electrophoresis of the PCR with XYN2 primers (B) Gel electrophoresis of PCR with promoter specific primer and XYN2 right primer