Telomerase/Telomere Biochemistry Overview
advertisement

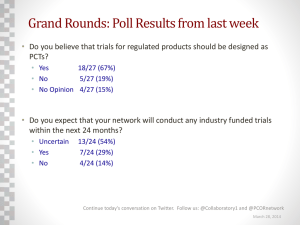
Mapping Protein-Protein Interactions MEDG 505 (Genome Analysis) 13 January 2005 •Morin: -Overview -IP-MS -Data integration •Student presentations: -Y2H interactions -RNA vs Protein expression analysis •Discussion: -Lessons -Application Central Dogma DNA RNA Protein Humans: - ~25,000 genes - 25-40% with functional annotations General Goal: Annotation of proteome -Identify disease related proteins -Identify therapeutic targets How identify protein functions? Function Protein Function General purpose of proteins is to interaction with other molecules -Enzyme/substrate -Protein/protein Cellular processes governed by complex networks of interacting proteins -Determination of protein-protein interactions infers functional hypotheses Protein Annotation Large Scale Methods for annotation of protein function: -Genetic -can verify biological role -Mutational analysis in model organisms -verifies biological role -binary interactions -Yeast 2-hybrid -comprehensive and HTP -translation to humans -often protein fragments -Genomic -mRNAs infer proteome problematic -high false positives -mRNA profiling -identifies expression -differences in biologychanges cloud -identifies interactions -extensively employed directly -Biochemical -silent to PTMsorder interactions interpretation -yields higher -MS analysis of purified protein complexes -cause and PTMs effect difficult to infer -identifies -interactions difficult to employed predict -binding affinity can be -technically challenging Lesson: All methods need to be employed to fully annotate proteome. IP-MS Immunoprecipitation Mass Spectrometry Immunoprecipitate Interaction Partners Protein identification Gel separation Excise bands LC-MS/MS fragmentation Tagged Protein Structure N-tagged construct CMV FLAG lox lox ORF C-tagged construct CMV lox ORF lox FLAG Properties of Immunoprecipitated Protein Complexes Types of interacting proteins • Background binding to bait/matrix/MS (filter?) • Proteins from throughout lifespan • Processing/transport/degradation proteins (filter?) • Weak affinity (less reproducible?) • Strong affinity • Primary interactors • Secondary interactors • High data volume Experimental design and analysis should be designed for expectations Methodology for evaluation 1-Experimental validation 2-Bioinformatic evaluation 3-Experimental reproducibility -transfection/IP protocols Method Characterization Characterization Project 1- 49 Baits, from diverse protein families -tag both N and C termini 2- IP-MS, repeat 4+ times 3- 190 preys -hit: -observed 2+ times -frequency less than 5% 4- Analyze N- & C-Tag Hit Overlap seen with N only seen with C only seen in both N&C seen when N+C are combined total # hits 110 29 15 8 162 % of total hits 0.68 0.18 0.09 0.05 Fraction of total hits observed N-tag only experiment 0.77 C-tag only experiment 0.27 Lessons: 1) 5 Hits per Bait. 2) N-tags interfere less than C-tags. 3) Both tags needed to get good representation. Sample 33 Baits Prey Reproducibility Observed Reproducibility Rate 0.39 0.31 0.40 0.30 N 0.17 Average C 0.04 0.01 0.07 0.10 0.02 0.20 0.01 Fraction of Hits 0.50 0.00 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 Reproducibility Rate Sample 42 Baits 190 Preys Note: ~50% of C-tags have 1.0 rate. Lesson: Improve immunoprecipitation conditions. Question: How many trials to see a prey 2 times? Planning Trial Size Number of Trials Needed to Observe Prey 2+ Times 1 0.9 Fraction of Hit Pool 0.8 # of trials 0.7 2 3 0.6 4 0.5 5 0.4 6 0.3 Reproducibility Rate 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 Theoretical Probability of 2+ observations in X # of trials 2 3 4 5 6 0.00 0.00 0.00 0.00 0.00 0.01 0.03 0.05 0.08 0.11 0.04 0.10 0.18 0.26 0.34 0.09 0.22 0.35 0.47 0.58 0.16 0.35 0.52 0.66 0.77 0.25 0.50 0.69 0.81 0.89 0.36 0.65 0.82 0.91 0.96 0.49 0.78 0.92 0.97 0.99 0.64 0.90 0.97 0.99 1.00 0.81 0.97 1.00 1.00 1.00 1.00 1.00 1.00 1.00 1.00 0.2 0.1 0 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0 Reproducibility Rate Binomial distribution equation n! n Lessons: (Lessons: Pobs) k ( p k )((1 p) nk ) ksuspect !(n k )! •Identifies •Identifiessuspect data data p: prey observation frequencyrate •Improving •Improving reproducibility reproducibility rate reduces number oftrials trialsneeded. needed. n:reduces number number of trials of k: number of observations required (2) Fraction Predicted Fraction of Observed = 0.5Prey Pool Found in X # of trials ReproducibilityRate of Prey 2 3 4 5 6 2 Rate trials 0 Pool0.00 0.003 trials 0.00 0.00 0.00 0.00 H 0.00 T0.00H0.00H 0.00 H H 0.1 H 0.00H 0.00 0.2 0.01 0.00 0.00 0.00 0.00 0.00 T 0.00 H0.01H0.01T 0.01 H T 0.3 T 0.02H 0.00 0.4 0.07 0.01 0.02 0.03 0.04 0.05 H 0.19 T0.27T0.31H 0.34 T H 0.5 H 0.39T 0.10 0.6 0.01 0.00 0.01 0.01 0.01 0.01 T 0.03 H0.04T0.04T 0.04 T T 0.7 T 0.04T 0.02 0.8 0.17 0.11 0.15 0.16 0.17 0.17 0.9 0.00 0.00 0.00 0.00 0.00 0.00 Note: 1 0.31 0.31 0.31 0.31 0.31 0.31 •If hit = 3+ times then 0.125 1.00 0.55probability 0.72 0.83 = 0.89 0.93 False Negative Rate Predicted False Negative Rate 1 Fraction of Hit Pool 0.9 0.8 0.7 0.6 0.5 0.4 0.3 0.2 Reproducibility Rate 0 0.1 0.2 0.3 # of trials 0.4 2 0.5 3 0.6 4 0.7 5 0.8 6 0.9 1 0.1 0 0.0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0 Reproducibility Rate Lesson: •1 or 2 trials provides highly incomplete dataset. Reproducibility Rate 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 1 Theoretical Probability of NOT Observing 2+ in X # of trials 2 3 4 5 6 1.00 1.00 1.00 1.00 1.00 0.99 0.97 0.95 0.92 0.89 0.96 0.90 0.82 0.74 0.66 0.91 0.78 0.65 0.53 0.42 0.84 0.65 0.48 0.34 0.23 0.75 0.50 0.31 0.19 0.11 0.64 0.35 0.18 0.09 0.04 0.51 0.22 0.08 0.03 0.01 0.36 0.10 0.03 0.01 0.00 0.19 0.03 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 Fraction of Prey Pool 0.00 0.00 0.01 0.02 0.07 0.39 0.01 0.04 0.17 0.00 0.31 1.00 Predicted Fraction of Prey Pool NOT Found in X # of trials 2 3 4 5 6 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.01 0.01 0.01 0.01 0.01 0.05 0.04 0.03 0.02 0.01 0.29 0.19 0.12 0.07 0.04 0.01 0.00 0.00 0.00 0.00 0.02 0.01 0.00 0.00 0.00 0.06 0.02 0.01 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.00 0.45 0.28 0.17 0.11 0.07 Predicted False Positive Rate Predicted False Postive Rate vs. Database Frequency 1 False positive frequency Method -determine prey frequency in database -Assume background proteins have a uniform random distribution -Assume background does not change with time or experimental conditions -Compare prey frequency to predicted observation rate < 0.05 0.9 0.8 0.7 0.6 0.5 # of trials 0.4 2 3 4 5 6 0.3 0.2 0.1 0 0 0.1 0.2 0.3 0.4 0.5 0.6 0.7 0.8 0.9 PathMap (global) observation frequency risk ( p) k 2 mn n! ( p k )((1 p) n k ) k!(n k )! E falsepositive (cutoff) p 0 p cutoff risk ( p) Numhits( p) p: prey observation frequency “safe” 5% n: number of trials region k: number of observations required (2) Efalsepositive: expected number of false positives cutoff: frequency cutoff Numhits(p): number of hits at each prey observation frequency 1 Estimated Experimental False Positive Rate Random Sampling Method -randomly reassign bait labels for each IP for all 49 baits -repeat -obtain 3, 4, and 5 trial sets, 49 baits each, with preys randomly assigned to a bait (5% database frequency) -assume random distribution (no relation between baits) Results # trials 3 4 5 Observed reproduced hits (false positives) percent Calculated number number (n=190) of false postives 1 6 7 1% 3% 4% <1 <2 <3 -false positive rate 2-3X greater than calculated. -non-uniform distribution Reasons -not independent experiments -non-random -baits are related -cross-contamination -equipment contamination Managing False Positives 1-Control subtraction -empty vector immunoprecipitation -irrelevant protein immunoprecipitation 2-Reproducibility -2+ times -3-4 biological replicates 3-Database frequency -observation frequency cutoff 4- Prioritization -annotation 5-Validation -reciprocal immunoprecipitation -co-expression Interaction Network Example Human Pathway Pilot Project TNFa pathway -Proinflammatory cytokine expressed mainly by activated monocytes and macrophages -Highly studied -Pathway members provide ready availability of baits. -Understanding incomplete, providing opportunity for discovery -Disease involvement -Tumor progression and killing -Diabetes -Infection -Inflammation -Pharmaceutical potential -Find protein targets that perform isolated TNFa functions without side-effects. Contract design: -20 baits, chosen by customer (17 actually provided) -N & C FLAG tags, constructed by MDSP. -Report all observed interactions. Additional design parameters: -Expressed and immunoprecipitated 4 times each. -Report all interactions classified as hits. TNFα Pathway: Inflammation/Cancer with Preys - 17 Baits - Both N & C tags - 4 Immunoprecipitations NK Cell Function TGFr IL10RB ENaC CD40 IL10RA Na Channels Fas Endocytosis Regulation KIAAxxx TNFr CS1 xxx4 Rab5 SGK xxx15 DNA repair/Damage xxx14, xxx2 TRAF2 xxxF1 xxx1 xxxCA TBK1 TANK xxx1-L Caspases NIK Stat xxxB Others xxxL1, xxxC1, FLJxxx, FLJxxx, xxx1, MGCxxx, KIAAxxx, FLJxxx, xxxA11 xxxA13 Xxxx xxxA3 IKAP Cell Death IKK-1 Transcriptional Regulation xxx8 FLJxxx IKB Src Protein Transport xxx13 NF-kB Ptyr PP xxx4, xxxA, xxxE, xxxG1, xxxG2, xxx4 xxx5 B-xxx1 xxx12 TRAF3 Jak xxx1A xxx37 TRAF6 xxxCB CLARP RIPK2 FADD Protein Sorting / Targeting xxx23, xxx-SR, xxx3, FLJxxx, xxx3, xxx4 xxxB12 xxx7 xxx19 KIAAxxx xxx8 Gxxx xxx-99 3-xxx 14-3-3 xxx1 PPP 14-3-3 xxx1D xxx xxx1 14-3-3 xxx1 14-3-3 xxx FLJxxx 14-3-3 xxx4 xxx PP1CB XRCC7 xxx11 GYS1 PPP1R3 xxxA1 Nucleus Transcription xxxL1 CDC2 xxx4 xxx xxx14 xxxG4 xxxL1 kinase A20 xxx130 SGK Gene xxxA' xxxA9 Transcription xxxB xxxA1 TNFa Bait Protein Other TNFa Pathway protein Prey protein Interactions with Bait protein ??? xxxA8 Activation Cyclin xxxGP KIAAxxx PITSLRE(8) ??? Cell cycle Control Transcription Inhibition Causal (indirect) interactions CS1/Jasmine/19A24 Gene TNFa Pathway Project Summary Bait information baits membrane baits expressed membrane baits expressed baits with interactions expressed baits with no TNFa context Potential antibody targets number comment 17 3 14 2 not expected to express 2 13 7 Bait/Prey information preys known interactions new interactions baits placed in context new bait/prey/bait linkages 99 13 86 5 4 also observed 1 known linkage Prey information enzymes 37 proteins in druggable families proteins with no function hypothetical proteins transmembrane (TM) domain containing proteins 7 TM potential plasma membrane proteins protease, GTPase, ATPase, kinase, 20+ phosphatase, receptor 13 6 enzymes, 1 receptor 4 15 1 receptor? 8 others ER or mitochondrial Integrating Proteomic and Genomic Information Genes Regulating Cell Growth and Division Systematic identification of pathways that couple cell growth and division in yeast Science 297: 395-400, 2002. Paul Jorgensen Joy L. Nishikawa Bobby-Joe Breitkreutz Mike Tyers Program in Molecular Biology and Cancer Samuel Lunenfeld Research Institute Mount Sinai Hospital Toronto, Ontario, Canada Genetic Screen for Yeast Size Mutants whi lge 4812 strains (~2 yrs) Wild type size profile sfp1 whi lge 10 35 60 85 Cell volume (fL) 110 SFP1 regulated genes WT GALSFP1 SFP1 GAL genes (10) Nucleotide biosynthesis (12) tRNA synthetases (6) ribosome biogenesis (21) RNA Polymerases I and III (10) nucleolus (29) Translation initiation and elongation (17) Ribosomal protein genes (136) scale 5 3 1.5 -1.5 -3 -5 Yeast Interaction Map Ho et al. Nature 10:180-3, 2002. aFLAG IP > LC-MS/MS -725 bait attempts -493 baits > 1578 preys -646 unannotated preys Overlap of Genetic, Expression & Interaction Data Protein interactions Genetic interaction Common mRNA regulation Nucleolar Network Gene Regulation in Breast Cancer 98 breast tumors x 25000 genes “genes that are overexpressed in tumors with a poor prognosis profile are potential targets for the rational development of new cancer drugs” 430 van’t Veer et al. (2002) Nature 415, 530-6. 2460 Proteins in the functional pathway of disease associated genes may identify additional or better 231 therapeutic targets. Overlap of PathMap and Breast Cancer Genes MDSP reporter ER BRCA1 Prognostic Rosetta 2460 430 231 van’t Veer et al. (2002) Nature 415, 530-6. primary enz 194 8% 42 27 6% 7 28 12% 9 secondary enz 515 87 208 38 27 4 Protein Networks in Prognosis Reporters enzyme + 55 only 35 up regulated 4 down regulated 16 Interaction network provides context Integrated Genomic/Proteomic Breast Cancer Project reporter # of genes ER BRCA1 Prognostic 2460 430 231 •Profile gene expression changes during tumor progression •Assemble experimental gene set -genes with expression changes -genes suspect for breast cancer progression •Perform IP-MS to determine interacting proteins •Analyze for regulatory networks and critical pathways van’t Veer et al. (2002) Nature 415, 530-6.