Figure S1 - Springer Static Content Server
advertisement

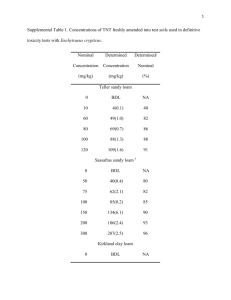
Supplementary Information: Supplementary Text Supplementary References Figure S1 Figure S2 Figure S3 Figure S4 Figure S5 Figure S6 Table S1 Table S2 Table S3 Supplementary Text: The frequency of gene coding DNA (cDNA) migration in relationship to geographic distance is shown on a plot of sample site GPS coordinates within YNP (Figure 1). YNP regions from Figure 1 are depicted in Figures S1 through S4, focusing in on several well sampled regions within the Park, including Lower Geyser Basin (Figure S1), Bison Pool (Figure S2), Norris Geyser Basin (Figure S3) and Mammoth Hot Springs (Figure S4). As with Figure 1, in Figures S1 through S4 sites are connected with lines demonstrating recent cDNA migration with line thickness indicates normalized migration quantity and thicker lines indicating higher quantities of DNA migration/sharing between sampled sites. Sites and connections are colored as phototrophic (blue) and chemotrophic (red), with three examples of chemotrophic to phototrophic cDNA migration in green (all three involve BP3, a ‘transitional’ chemotrophic community positioned just above the photosynthetic fringe in Bison Pool. Figures S1 through S4 are broken into four quadrants of increasing Ks ranges (Ks < 0.01 at top left, 0.01 <= Ks < 0.07 at top right, 0.07 <= Ks < 0.32 at bottom left and 0.32 <= Ks < 0.50 at bottom right. As reiterated in iterative multiple linear regression analysis (in main text), figures S1-S4 underscore that scaffolds are shared/migration can occur over long distances across YNP; geographic distance is not a primary factor in determining the frequency of DNA migration between sites. For instance, there is an absence of DNA migration between YNP 7 and neighboring sites (including YNP 3, 8, 14, and 19) despite these sites lying only a few kilometers apart, whereas DNA migration is occurring over 70+ km separating YNP 12 and YNP 13. Migration most commonly occurs between sites whose communities employ similar metabolic strategies, i.e. shared scaffolds are detected between phototrophic sites and chemotrophic sites, but seldom between the two. BP 3, mentioned above, is a noted exception, with low Ks scaffolds (arguably recent migrations) shared with BP 4, BP 5 and YNP 15 (all phototrophic sites. The most abundant DNA migrations occur between Aquificae-dominated alkaline environments, including YNP 11, YNP 12, YNP 13, BP1 and BP2. Concomitantly, the range of environments that host Aquificae span the entire geographic area of the Park. Additionally, strong links exist between acidic sites, including YNP 4, YNP 8, YNP 9 and YNP 18, which also cross the width of the Park. Interestingly, YNP 4 and YNP 12 are similar temperature (78oC vs 75oC, respectively) chemotrophic sites with a relatively small geographic separation (~15 km), yet these two communities show no shared DNA fragments—despite both being DNA migration ‘hubs’ to other communities, each sharing fragments with multiple other sites across the Park. This reiterates the idea that multiple environmental factors are involved in selecting for, maintaining, and also attenuating the biological connections between sites. Within the plots shown in Figures S1 through S4, the majority of DNA migrations in the lowest Ks quadrant (top left) are among phototrophic sites (blue lines) with migrations among chemotrophic sites (red lines) becoming prevalent in mid-Ks ranges (top right and lower left), until the highest Ks quadrant (lower right) is dominated by migrations among chemotrophic sites (red lines). These observations in essence represent the overall trends shown Figure 3 in the main text: phototroph communities appear to be sharing DNA more frequently across separate sites. Intriguingly, several shared scaffolds were detected between YNP and GBS (a distance of ~800km), though these are present only in the highest Ks quadrant, consistent with the large temporal and geological/geographical separation between GBS and YNP communities. Principal components analysis (PCA) was performed on 20 of the geochemical parameters measured across all YNP metagenomic sites (Figure S5A). This analysis resulted in two principal components (PC1 and PC2) accounting for 44.2% of the geochemical variation between sampled locations. A biplot of PC1 verses PC2 (Figure S5A) indicates the location of metagenomic sites in geochemical space with vectors indicating the directionality and magnitude of influence of each geochemical parameter. Sampled locations in similar regions of YNP are circled and tight clustering of geochemically and geographically distinct regions within YNP is clearly visible. The five samples taken along the Bison Pool outflow channel (within the Lower Geyser Basin grouping) demonstrate tight clustering despite a 32oC temperature gradient, confirming the PCA analysis is clustering across multiple geochemical dimensions that, for Bison Pool in particular, remain relatively constant along the outflow channel. Additionally, pH is a major vector along PC2, separating the acidic Norris Geyser Basin sites from the alkaline Lower Geyser Basin sites. Figure S5B maps DNA migration in this PCA space, using lines to represent DNA migration between sites sorted based on their physico-chemical similarities. Line thickness is proportional to the number of shared scaffolds between sites and lines are color-coded to highlight community metabolism, as described above. Notabl, proximal points in this PCA space tend to have the strongest connections; shared scaffolds are most frequently found between communities that occupy geochemically similar environments. This is especially evident with the three phototrophic-to-chemotrophic connections, all occupying a very small region of the PCA plot. Despite the geographic separation of ~10 km between BP3 and YNP 15, they are closer than BP3 and BP5 in geochemical space (~10m separation), further supporting the notion that “environment selects” what migrations are ultimately fixed in a community. YNP 4, a slightly acidic site with pH 6.1, is serving as a bridge with connections to both acidic sites (YNP 8 and YNP 14) and alkaline sites (YNP 11 and BP1). The Mammoth Hot Springs area is poorly connected to the rest of YNP, as is demonstrated by YNP 10 and YNP 18 exhibiting only weak connections despite being geochemically similar chemotrophic sites. While distance is frequently overshadowed by other factors in predicting numbers of shared scaffolds between sites, hydrothermal regions within the Park do appear in this PCA space (circled areas in Figure S5), and the hot springs within these regions do show broad geochemical similarities (Table S1). However, YNP 13 is ~25 km away from BP1 and yet YNP 13 and BP1 demonstrate the largest quantity of high identity scaffolds shared between any pair of sites across the Park. The PCA analysis also accounts for the lack of migration between YNP 4 and YNP 18, which are less than 10 km apart but reside on opposite sides of the PCA plot, with YNP 4 clustering near the Lower Geyser Basin sites and YNP 18 plotting closer to the Mammoth Hot Springs sites in Figure S5. To investigate biases in the phyla responsible for DNA migration between sites, 2-way clustering was done using all scaffolds in each metagenomic site (Figure 2) and the scaffolds involved in DNA migration (Figure S7B). Phyla were assigned as the best hit within the NCBI nt database, if one existed, for all scaffolds over 3,000bp in length. The taxonomic family of all cDNA fragments involved in DNA migration was determined to investigate bias in Ks distribution and migration frequency. Figure 2 contains a bar plot which demonstrates the fraction of all cDNA migrations assigned to each taxonomic family, including the fraction of unclassified cDNA sequences transferred between chemotrophic (top) and phototrophic (bottom) sites. Families are ordered and colored by their distribution within chemotrophic (top - red) and phototrophic (bottom - blue) sites, with families found within both phototrophic and chemotrophic communities colored grey. Additionally, a Ks histogram was generated for each taxonomic family between Ks values of 0.00 and 0.50 (right of family name). The largest percentage of cDNA migrations are attributed to unclassified cDNAs from chemotrophic sites (25%), with Aquificacaea (25%) and Chloroflexaceae (10%) accounting for the highest percentages of chemotrophic and phototrophic transfers. Interestingly, none of the families commonly found within both chemotrophic and phototrophic sites (grey bars) accounted for more than 1% of cDNA transfers, indicating a strong bias towards the migration of families limited to either high temperature (chemotrophic) sites or photosynthetic families. The Ks histograms for highly transferred families mirror the overall trends seen in Figure 1, for instance with the Aquificacaea histogram representing a Poisson distribution centered at 0.18 and, similarity, Acidobacteriacea, Chloroflexaceae and Synococcacaea (all known photosynthetic phyla) demonstrating large peaks at very small (<0.01) Ks values. Supplementary References: 35. I. V. Grigoriev et al., The Genome Portal of the Department of Energy Joint Genome Institute, Nucleic Acids Res. (2011), doi:10.1093/nar/gkr947. 36. S. F. Altschul, W. Gish, W. Miller, E. W. Myers, D. J. Lipman, Basic local alignment search tool, J. Mol. Biol. 215, 403–410 (1990). 37. M. Kanehisa, S. Goto, KEGG: Kyoto Encyclopedia of Genes and Genomes, Nucleic Acids Res. 28, 27–30 (2000). 38. Z. Zhang et al., KaKs_Calculator: calculating Ka and Ks through model selection and model averaging, Genomics Proteomics Bioinformatics 4, 259–263 (2006). Figure S1: Figure S2: Figure S3: Figure S4: Figure S5: Table S1: YNP environmental metadata for 24 YNP metagenomes used in this study, listed by sample site shorthand and hot spring name (bdl=concentrations below detection limits, mM=millimolar aqueous concentrations, ionization/speciation given if known) T pH °C Na+ mM K+ mM Al mM Fe mM Mg mM ClmM NH4+ SO4-2 NO3- mM mM mM mM P mM Si mM B mM As mM Zn mM Mn mM S2mM O2 mM 425 0.04 685 8.74 bdl 5.2 5.36 0.11 2.0 3.1 10.2 bdl bdl Ca+2 Alice Springs 75 2.1 1.1 0.38 0.68 396 227 YNP2 Nymph Lake 89 4.7 4.6 0.29 0.39 37.4 1.5 57 4.1 230 1.34 5.8 0.11 2.82 0.26 8.7 1.3 33.9 3 bdl YNP3 Monarch Geyser 85 4 9.5 0.64 0.08 29.6 3.6 8.6 9.7 277 1.33 12 bdl 5.29 0.58 19.4 1.4 1.3 7.8 bdl YNP4 Joseph's Coat 78 6 10.5 2.00 0.42 3.0 1.6 43.6 8.9 8424 3.79 12 bdl 5.58 5.85 165 bdl 5.3 20 bdl YNP5 Bath Lake 57 6.2 3.93 0.97 8.82 bdl 0.7 2132 4.4 40 5.60 4.8 87.7 0.66 0.31 24 0.3 0.2 117 bdl YNP6 White Creek 52 8.2 3.59 0.39 0.42 3.3 1.7 13.8 1.8 1.9 0.23 2.1 3.2 3.83 0.07 5 0.2 4.7 bdl 188 YNP7 52 5.8 4.12 0.44 0.47 2.7 75.5 58.8 0.89 4.2 0.23 2.2 54.9 1.38 0.04 9 bdl 24 bdl bdl 73 3.3 10.8 0.28 0.11 64.8 36.6 14 14.2 88 1.05 19 bdl 10.4 0.69 28.4 1.5 1.6 10 22 YNP10 Chocolate Pots One Hundred Springs Plain Narrow Gauge 71 6.5 4.42 1.10 8.40 0.7 0.4 2219 4.55 40.1 5.66 6.5 38 0.72 0.30 20 0.0 0.4 67 bdl YNP11 Octopus Spring 82 7.9 12 0.34 0.01 8.1 0.2 bdl 6.60 2.3 0.14 6.0 9 6.11 0.24 27.0 0.0 bdl 0 34 YNP12 Calcite Spring 75 7.8 8.81 2.40 0.60 8.1 3.2 251 6.62 1660 3.41 9.4 bdl 3.40 3.09 21 0.2 1 105 bdl YNP13 81 7.8 9.70 0.34 0.10 1.7 bdl 1 0.22 bdl bdl 2.90 bdl 2 0.3 1 bdl bdl 73 3.5 10.8 1.38 0.10 65.3 31.6 9.1 87 1.18 18.5 bdl 5.67 0.65 24 1.0 1.2 3 30 59.9 8.2 12.6 0.64 0.02 2.9 0.1 0.8 2.00 13.2 0 7.30 bdl YNP15 Bechler One Hundred Springs Plain Mushroom Spring 4.4 0.18 6.6 bdl 4.51 0.25 26 0.5 0.1 bdl 141 YNP16 Fairy Geyser 35 8.1 9.38 0.10 0.05 3.1 bdl 3.5 5.20 1.3 0.18 6.7 11.9 0.21 13 bdl bdl bdl 31 YNP17 Obsidian Pool Prime 56 5.8 12.4 0.85 0.13 9.6 3.1 24.7 8.40 5.9 bdl 2.38 10.0 0 0.60 8 0.5 1 bdl 60 46.0 23 1.82 0.55 3.9 0.0 bdl 157 bdl bdl bdl 6.70 0.77 18.8 6.4 bdl 16.2 bdl YNP1 YNP8 YNP14 YNP18 Washburn Spring 76 6.4 1.2 0.28 0.59 bdl bdl 451 0.04 YNP19 Cistern Spring 76 4.4 11.3 0.97 0.05 8.2 0.2 3 14 0.6 2.50 2396 0 296 16.6 7 0.74 YNP20 Bath Lake 54 6.2 5.05 1.25 9.75 6.7 0.7 2625 5.70 40 7.20 5.6 bdl 0.70 0.30 23 0.3 bdl 117 bdl BP1 Bison Pool Site 1 93.3 7.35 12.9 0.26 0.02 19.2 bdl 2.72 6.11 bdl 0.21 50.0 3.07 4.58 0.22 11.8 0.05 bdl 4.77 5.4 BP2 Bison Pool Site 2 79.4 7.68 13.2 0.26 0.02 19.6 bdl 2.57 6.29 bdl 0.20 50.0 2.80 6.34 0.24 12.3 0.06 0.01 2.03 24 BP3 Bison Pool Site 3 67.5 7.92 13.7 0.28 0.02 15.3 0.02 1.11 6.48 bdl 0.20 35.7 bdl 6.65 0.26 14.9 bdl bdl 3.12 28 BP4 Bison Pool Site 4 65.3 8.00 13.8 0.28 0.02 29.1 bdl 2.51 6.52 bdl 0.20 42.8 3.64 5.80 0.31 15.8 0.03 0.01 4.68 50 BP5 Bison Pool Site 5 57.1 8.26 13.8 0.28 0.02 19.1 0.02 bdl 6.60 bdl 0.19 42.8 1.57 5.80 0.26 13.4 0.01 bdl 2.18 87.5 Table S2: Multiple Regression Results All Metadata CoefficientStd. Error Intercept NO3 K SO4 t value p value 1133.37 349.39 3.24 0.002 13.32 6.53 2.04 0.04 -610.94 320.50 -1.91 0.06 -67.51 43.90 -1.54 0.13 Zn -126.60 82.58 -1.53 0.13 pH -156.78 106.25 -1.48 0.14 O2 -3.61 2.53 -1.43 0.16 T -18.47 13.60 -1.36 0.18 As -7.64 5.82 -1.31 0.19 B 259.51 199.81 1.30 0.20 Na -58.87 47.39 -1.24 0.22 GPS 1.65 1.87 0.88 0.38 Si 64.65 80.41 0.80 0.42 Cl 34.86 52.85 0.66 0.51 S 2.64 4.35 0.61 0.55 Al 1.99 4.42 0.45 0.65 P -2.14 5.59 -0.38 0.70 Ca -70.58 248.99 -0.28 0.78 Mg 0.27 1.00 0.27 0.79 Fe -1.54 7.24 -0.21 0.83 Mn -2.66 16.26 -0.16 0.87 Adjusted R-squared: 0.04, p-value: 0.25 After Iterative Multiple Linear Regression (iMLR) t value p value Intercept CoefficientStd. Error 1073.46 281.32 3.82 0.0002 K -468.00 190.21 -2.46 0.02 12.08 5.56 2.17 0.03 -159.90 78.23 -2.04 0.04 SO4 -48.26 24.95 -1.93 0.06 T -19.28 12.34 -1.56 0.12 B 238.30 154.31 1.54 0.13 As -7.48 4.89 -1.53 0.13 O2 -3.23 2.25 -1.44 0.15 NO3 pH Adjusted R-squared: 0.10, p-value: 0.01 Table S3: Over/under-representation of enzymes in shared metagenome fragments (EC enzyme commission #; K# KEGG orthology #; bold+italics p<.01; italics p<.05) EC Annotation Observed Total 75 Observed Shared Fragments 175 Expected Shared Fragments 49 log2 odds -1.84 pvalue 0.0002 4.1.1.61 K01612 4-hydroxybenzoate decarboxylase 6.4.1.1 biotin carboxylase 371 930 254 -1.87 0.0002 1.8.2.1 sulfite dehydrogenase SoxC precursor 30 55 17 -1.69 0.0024 1.4.3.19 K03153 glycine oxidase 125 211 66 -1.68 0.0028 1.3.3.1 K00226 dihydroorotate oxidase 355 6 71 3.56 0.0037 3.6.3.25 K02045 sulfate transport system ATP-binding protein 721 13 143 3.46 0.0041 2.6.1.42 K00826 branched-chain amino acid aminotransferase 330 7 66 3.24 0.0076 3.6.3.30 K02010 iron(III) transport system ATP-binding protein 1133 24 226 3.24 0.0076 2.6.1.62 DapA aminotransferase 127 172 58 -1.57 0.0097 2.4.1.83 K00721 dolichol-phosphate mannosyltransferase 536 6 106 4.14 0.0103 1.8.1.9 K00384 thioredoxin reductase (NADPH) 530 12 106 3.14 0.0106 5.4.99.12 K03177 tRNA pseudouridine synthase B 989 10 195 4.29 0.013 6.3.4.14 340 430 150 -1.52 0.0141 238 6 48 3 0.0162 2.7.7.42 K11263 acetyl-/propionyl-CoA carboxylase, biotin carboxylase, biotin carboxyl carrier protein K02013 iron complex transport system ATP-binding protein putative glutamate-ammonia-ligase adenylyltransferase 143 171 61 -1.49 0.017 3.1.1.29 K01056 peptidyl-tRNA hydrolase, PTH1 family 313 8 63 2.98 0.0172 2.7.1.2 K00845 glucokinase 274 7 55 2.97 0.0177 2.4.1.227 194 220 81 -1.44 0.0225 292 8 59 2.88 0.0226 8 8 3 -1.42 0.0249 1.1.1.3 K02563 UDP-N-acetylglucosamine--N-acetylmuramyl(pentapeptide) pyrophosphoryl-undecaprenol Nacetylglucosamine transferase K00548 5-methyltetrahydrofolate--homocysteine methyltransferase K03280 UDP-N-acetylglucosamine:(glucosyl)LPS alpha-1,2-N-acetylglucosaminyltransferase K00003 homoserine dehydrogenase 336 10 68 2.77 0.0298 6.4.1.3 K01966 propionyl-CoA carboxylase beta chain 268 8 54 2.75 0.0312 4.1.3.27 K01657 anthranilate synthase component I 689 21 139 2.73 0.0327 3.6.3.34 2.1.1.13 2.4.1.56 2.5.1.72 K03517 quinolinate synthase 229 7 46 2.72 0.0335 6.6.1.1 K03403 magnesium chelatase subunit H 456 14 92 2.72 0.0335 6.1.1.24 glutamyl- and glutaminyl-tRNA synthetase 78 79 31 -1.35 0.034 1.1.1.132 K00066 GDP-mannose 6-dehydrogenase 32 33 13 -1.34 0.0354 1.3.3.4 K00231 protoporphyrinogen oxidase 127 124 49 -1.34 0.0354 5.4.3.2 lysine 2,3-aminomutase 122 119 47 -1.34 0.0354 1.1.1.267 1-deoxy-D-xylulose 5-phosphate reductoisomerase 209 200 80 -1.32 0.0382 2.7.2.11 K00931 glutamate 5-kinase 173 161 65 -1.31 0.0397 3.1.13.1 K01147 exoribonuclease II 95 86 35 -1.3 0.0412 4.2.1.46 K01710 dTDP-glucose 4,6-dehydratase 633 21 128 2.61 0.0426 2.6.1.85 K03342 para-aminobenzoate synthetase / 4-amino-4deoxychorismate lyase K01586 diaminopimelate decarboxylase 144 131 54 -1.28 0.0443 257 221 93 -1.25 0.049 253 9 51 2.5 0.0531 1.2.7.8 K00166 2-oxoisovalerate dehydrogenase E1 component, alpha subunit indolepyruvate oxidoreductase subunit IorB 251 9 51 2.5 0.0531 4.1.99.3 K01669 deoxyribodipyrimidine photo-lyase 361 300 129 -1.22 0.054 5.4.99.5 K01850 chorismate mutase 202 163 71 -1.2 0.0574 3.1.1.61 K03412 two-component system, chemotaxis family, response regulator CheB K10206 LL-diaminopimelate aminotransferase 624 23 126 2.45 0.0583 223 179 79 -1.18 0.061 K06044 (1->4)-alpha-D-glucan 1-alpha-Dglucosylmutase K01626 3-deoxy-7-phosphoheptulonate synthase 159 125 55 -1.18 0.061 262 10 53 2.41 0.0627 K01703 3-isopropylmalate/(R)-2-methylmalate dehydratase large subunit K01537 Ca2+-transporting ATPase 617 475 213 -1.16 0.0646 413 16 84 2.39 0.065 233 9 47 2.38 0.0662 4.2.1.17 K00965 UDPglucose--hexose-1-phosphate uridylyltransferase K01692 enoyl-CoA hydratase 331 13 67 2.37 0.0673 2.4.1.182 K00748 lipid-A-disaccharide synthase 141 104 48 -1.12 0.072 1.1.1.91 K05882 aryl-alcohol dehydrogenase (NADP+) 147 6 30 2.32 0.0733 4.1.1.20 1.2.4.4 2.6.1.83 5.4.99.15 2.5.1.54 4.2.1.33 3.6.3.8 2.7.7.12 150 6 30 2.32 0.0733 2.7.8.13 K00996 undecaprenyl-phosphate galactose phosphotransferase phospho-N-acetylmuramoyl-pentapeptide-transferase 200 145 67 -1.11 0.0739 1.1.1.86 K00053 ketol-acid reductoisomerase 262 188 88 -1.1 0.0758 2.8.1.6 K01012 biotin synthetase 281 203 95 -1.1 0.0758 1.2.99.2 697 29 142 2.29 0.0771 3.5.2.6 K03519 carbon-monoxide dehydrogenase medium subunit K01467 beta-lactamase 142 6 29 2.27 0.0797 1.6.99.3 K03885 NADH dehydrogenase 253 178 84 -1.08 0.0798 6.3.2.8 K01924 UDP-N-acetylmuramate--alanine ligase 284 201 95 -1.08 0.0798 3.6.3.17 542 23 110 2.26 0.081 2.4.1.18 K02056 simple sugar transport system ATP-binding protein K00700 1,4-alpha-glucan branching enzyme 197 136 65 -1.07 0.0818 1.7.99.4 K00372 nitrate reductase catalytic subunit 187 8 38 2.25 0.0823 2.8.3.16 K07749 formyl-CoA transferase 184 8 38 2.25 0.0823 2.5.1.90 K02523 octaprenyl-diphosphate synthase 164 7 33 2.24 0.0836 3.1.26.12 134 92 44 -1.06 0.0838 4.2.1.51 fused ribonucleaseE: endoribonuclease/RNA-binding protein/RNA degradosome binding protein K04518 prephenate dehydratase 234 161 77 -1.06 0.0838 6.1.1.2 K01867 tryptophanyl-tRNA synthetase 424 286 139 -1.04 0.0878 6.2.1.5 succinyl-CoA synthetase subunit beta 660 444 216 -1.04 0.0878 3.13.1.1 K06118 UDP-sulfoquinovose synthase 69 47 23 -1.03 0.0899 3.2.1.1 K01176 alpha-amylase 202 9 41 2.19 0.0904 4.1.2.4 K01619 deoxyribose-phosphate aldolase 198 9 40 2.15 0.0961 1.1.1.22 K00012 UDPglucose 6-dehydrogenase 485 309 155 -1 0.0962 1.13.12.16 K00459 nitronate monooxygenase 99 64 32 -1 0.0962 3.2.1.35 K01197 hyaluronoglucosaminidase 13 8 4 -1 0.0962 6.1.1.16 K01883 cysteinyl-tRNA synthetase 480 311 155 -1 0.0962 1.1.1.49 K00036 glucose-6-phosphate 1-dehydrogenase 193 122 62 -0.98 0.1005 1.20.98.1 K08356 arsenite oxidase large subunit 116 73 37 -0.98 0.1005 2.7.8.6 4.2.1.3 K01681 aconitate hydratase 1 541 340 172 -0.98 0.1005 4.2.1.9 K01687 dihydroxy-acid dehydratase 356 219 112 -0.97 0.1027 6.1.1.15 K01881 prolyl-tRNA synthetase 504 312 159 -0.97 0.1027 1.2.1.12 K00134 glyceraldehyde 3-phosphate dehydrogenase 333 16 68 2.09 0.1049 6.2.1.30 K01912 phenylacetate-CoA ligase 168 8 34 2.09 0.1049 1.17.1.2 4-hydroxy-3-methylbut-2-enyl diphosphate reductase 305 15 63 2.07 0.108 2.4.2.18 anthranilate phosphoribosyltransferase 242 12 50 2.06 0.1095 2.5.1.7 243 12 50 2.06 0.1095 4.99.1.1 K00790 UDP-N-acetylglucosamine 1carboxyvinyltransferase K01772 ferrochelatase 144 7 29 2.05 0.1111 4.2.1.11 phosphopyruvate hydratase 451 269 141 -0.93 0.1115 2.4.1.119 163 8 33 2.04 0.1126 286 166 88 -0.92 0.1138 2.1.1.35 K07151 dolichyl-diphosphooligosaccharide--protein glycosyltransferase K02495 oxygen-independent coproporphyrinogen III oxidase K00557 tRNA (uracil-5-)-methyltransferase 83 47 25 -0.91 0.116 3.3.1.1 K01251 adenosylhomocysteinase 359 208 111 -0.91 0.116 2.3.1.47 8-amino-7-oxononanoate synthase 141 82 44 -0.9 0.1183 2.7.7.4 K00958 sulfate adenylyltransferase 215 11 44 2 0.119 3.1.11.6 K03602 exodeoxyribonuclease VII small subunit 116 6 24 2 0.119 1.2.1.41 gamma-glutamyl phosphate reductase 201 114 62 -0.88 0.1229 2.1.1.32 LOC100276957 146 83 45 -0.88 0.1229 2.5.1.6 K00789 S-adenosylmethionine synthetase 447 248 136 -0.87 0.1253 2.6.1.17 K14267 N-succinyldiaminopimelate aminotransferase 251 139 76 -0.87 0.1253 3.5.1.98 K06067 histone deacetylase 1/2 110 62 34 -0.87 0.1253 3.6.1.1 K01507 inorganic pyrophosphatase 513 27 105 1.96 0.1256 3.5.1.28 K01448 N-acetylmuramoyl-L-alanine amidase 151 8 31 1.95 0.1273 3.6.5.3 K02355 elongation factor EF-G 1308 708 394 -0.85 0.13 1.2.4.1 K00163 pyruvate dehydrogenase E1 component 462 25 95 1.93 0.1306 3.1.2.6 K01069 hydroxyacylglutathione hydrolase 183 10 38 1.93 0.1306 1.3.99.22 1.4.7.1 K00284 glutamate synthase (ferredoxin) 164 9 34 1.92 0.1323 2.7.1.71 K00891 shikimate kinase 163 9 34 1.92 0.1323 1.2.7.3 1006 55 207 1.91 0.1341 1.97.1.4 K00175 2-oxoglutarate ferredoxin oxidoreductase subunit beta K04069 pyruvate formate lyase activating enzyme 693 38 143 1.91 0.1341 6.2.1.12 K01904 4-coumarate--CoA ligase 31 16 9 -0.83 0.1348 3.2.1.23 K12308 beta-galactosidase 199 11 41 1.9 0.1358 6.1.1.10 K01874 methionyl-tRNA synthetase 498 262 148 -0.82 0.1372 6.4.1.4 127 67 38 -0.82 0.1372 126 7 26 1.89 0.1375 5.1.1.4 K01968 3-methylcrotonyl-CoA carboxylase alpha subunit K00658 2-oxoglutarate dehydrogenase E2 component (dihydrolipoamide succinyltransferase) K01777 proline racemase 27 14 8 -0.81 0.1396 1.2.7.5 K03738 aldehyde:ferredoxin oxidoreductase 548 31 113 1.87 0.1411 4.6.1.1 K01768 adenylate cyclase 245 14 51 1.87 0.1411 4.3.2.2 K01756 adenylosuccinate lyase 379 195 112 -0.8 0.142 6.4.1.2 714 370 212 -0.8 0.142 2.7.6.5 K01961 acetyl-CoA carboxylase, biotin carboxylase subunit K00951 GTP pyrophosphokinase 304 156 90 -0.79 0.1445 2.7.7.22 K00971 mannose-1-phosphate guanylyltransferase 164 85 49 -0.79 0.1445 1.3.1.74 K08070 2-alkenal reductase 989 499 291 -0.78 0.1469 2.5.1.77 FO synthase subunit 1 196 98 57 -0.78 0.1469 2.5.1.48 K01739 cystathionine gamma-synthase 153 9 32 1.83 0.1483 1.5.99.8 K00318 proline dehydrogenase 100 6 21 1.81 0.1519 2.1.2.3 335 164 97 -0.76 0.1519 2.3.3.9 bifunctional phosphoribosylaminoimidazolecarboxamide formyltransferase/IMP cyclohydrolase K01638 malate synthase 136 8 28 1.81 0.1519 3.1.21.3 K01154 type I restriction enzyme, S subunit 744 44 154 1.81 0.1519 3.5.4.10 K11176 IMP cyclohydrolase 334 164 97 -0.76 0.1519 3.6.1.11 K01524 exopolyphosphatase / guanosine-5'triphosphate,3'-diphosphate pyrophosphatase K00963 UTP--glucose-1-phosphate uridylyltransferase 171 10 35 1.81 0.1519 306 150 89 -0.75 0.1544 2.3.1.61 2.7.7.9 3.4.24.64 K01412 mitochondrial processing peptidase 272 135 80 -0.75 0.1544 1.1.2.4 K00102 D-lactate dehydrogenase (cytochrome) 182 11 38 1.79 0.1557 2.3.1.12 K00627 pyruvate dehydrogenase E2 component (dihydrolipoamide acetyltransferase) K00363 nitrite reductase (NAD(P)H) small subunit 186 11 38 1.79 0.1557 83 40 24 -0.74 0.1569 195 95 57 -0.74 0.1569 1.9.3.1 K00620 glutamate N-acetyltransferase / amino-acid Nacetyltransferase K02267 cytochrome c oxidase subunit VIb 789 48 164 1.77 0.1594 1.12.1.2 NAD-reducing hydrogenase, alpha subunit 237 111 68 -0.71 0.1646 5.4.3.8 K01845 glutamate-1-semialdehyde 2,1-aminomutase 399 185 114 -0.7 0.1671 4.2.1.24 K01698 porphobilinogen synthase 251 16 52 1.7 0.173 2.5.1.1 K00787 farnesyl diphosphate synthase 266 17 55 1.69 0.1749 4.2.3.1 K01733 threonine synthase 588 265 167 -0.67 0.1749 2.2.1.1 K00615 transketolase 628 41 131 1.68 0.1769 2.4.2.14 K00764 amidophosphoribosyltransferase 355 158 100 -0.66 0.1775 2.6.1.1 K00812 aspartate aminotransferase 497 221 140 -0.66 0.1775 2.7.2.4 K00928 aspartate kinase 363 162 103 -0.65 0.1801 3.4.21.26 K01322 prolyl oligopeptidase 89 6 19 1.66 0.1809 3.5.2.3 K01465 dihydroorotase 442 193 124 -0.64 0.1828 4.1.1.36 phosphopantothenoylcysteine decarboxylase 296 129 83 -0.64 0.1828 6.3.2.5 K01922 phosphopantothenate-cysteine ligase 295 129 83 -0.64 0.1828 5.3.1.5 K01805 xylose isomerase 72 31 20 -0.63 0.1854 1.1.3.15 K00104 glycolate oxidase 395 169 110 -0.62 0.1881 3.1.3.11 fructose 1,6-bisphosphatase II 440 189 123 -0.62 0.1881 1.5.1.12 K00294 1-pyrroline-5-carboxylate dehydrogenase 177 12 37 1.62 0.1891 2.7.9.2 K01007 pyruvate, water dikinase 398 27 83 1.62 0.1891 2.3.3.13 K01649 2-isopropylmalate synthase 629 267 175 -0.61 0.1908 3.5.4.25 K01497 GTP cyclohydrolase II 242 104 68 -0.61 0.1908 6.3.4.18 K01589 5-(carboxyamino)imidazole ribonucleotide synthase 237 101 66 -0.61 0.1908 1.7.1.4 2.3.1.35 3.6.3.4 K01533 Cu2+-exporting ATPase 495 34 103 1.6 0.1933 6.3.5.5 K01955 carbamoyl-phosphate synthase large subunit 1041 437 289 -0.6 0.1935 2.6.1.11 359 151 100 -0.59 0.1961 1.1.1.271 K00821 acetylornithine/N-succinyldiaminopimelate aminotransferase K02377 GDP-L-fucose synthase 87 6 18 1.58 0.1975 2.7.11.1 serine/threonine kinase 35 1044 73 218 1.58 0.1975 3.6.1.19 K02428 nucleoside-triphosphate pyrophosphatase 172 12 36 1.58 0.1975 3.6.3.6 K01535 H+-transporting ATPase 87 6 18 1.58 0.1975 4.1.1.32 K01596 phosphoenolpyruvate carboxykinase (GTP) 145 10 30 1.58 0.1975 5.4.2.10 phosphoglucosamine mutase 88 6 18 1.58 0.1975 1.1.1.202 1,3 propanediol dehydrogenase 15 6 4 -0.58 0.1989 1.6.5.5 K00344 NADPH2:quinone reductase 209 85 57 -0.58 0.1989 4.1.1.17 K01581 ornithine decarboxylase 78 33 22 -0.58 0.1989 6.2.1.34 K12508 feruloyl-CoA synthase 16 6 4 -0.58 0.1989 1.4.3.16 K00278 L-aspartate oxidase 246 101 68 -0.57 0.2016 3.1.3.1 K01077 alkaline phosphatase 250 102 69 -0.56 0.2043 4.4.1.1 K01758 cystathionine gamma-lyase 78 31 21 -0.56 0.2043 3.1.3.5 stationary phase survival protein SurE 374 27 78 1.53 0.2082 2.1.2.2 K11175 phosphoribosylglycinamide formyltransferase 1 204 15 43 1.52 0.2103 2.7.7.24 K00973 glucose-1-phosphate thymidylyltransferase 504 37 106 1.52 0.2103 6.2.1.13 CoA-binding domain-containing protein 316 23 66 1.52 0.2103 2.7.7.13 K00966 mannose-1-phosphate guanylyltransferase 354 26 74 1.51 0.2125 3.6.3.21 K02028 polar amino acid transport system ATP-binding protein K01928 UDP-N-acetylmuramoylalanyl-D-glutamate-2,6-diaminopimelate ligase K01940 argininosuccinate synthase 287 21 60 1.51 0.2125 274 20 57 1.51 0.2125 333 131 91 -0.53 0.2125 K14469 acrylyl-CoA reductase (NADPH) / 3hydroxypropionyl-CoA dehydratase / 3hydroxypropionyl-CoA synthetase K05903 NADH dehydrogenase (quinone) 82 6 17 1.5 0.2147 79 6 17 1.5 0.2147 6.3.2.13 6.3.4.5 1.3.1.84 1.6.99.5 3.4.21.53 K01338 ATP-dependent Lon protease 701 52 147 1.5 0.2147 6.2.1.1 K01895 acetyl-CoA synthetase 765 57 161 1.5 0.2147 1.18.6.1 K11333 chlorophyllide reductase iron protein subunit X 146 11 31 1.49 0.2169 2.8.1.7 K11717 cysteine desulfurase / selenocysteine lyase 537 208 146 -0.51 0.2181 6.1.1.21 K01892 histidyl-tRNA synthetase 401 155 109 -0.51 0.2181 3.2.1.21 K05349 beta-glucosidase 387 29 81 1.48 0.2191 3.4.11.9 K01262 Xaa-Pro aminopeptidase 240 18 50 1.47 0.2213 1.12.99.6 K06281 hydrogenase large subunit 197 15 41 1.45 0.2258 2.7.1.49 249 19 52 1.45 0.2258 3.6.3.3 K00941 hydroxymethylpyrimidine/phosphomethylpyrimidine kinase K01534 Cd2+/Zn2+-exporting ATPase 144 11 30 1.45 0.2258 2.7.1.17 K00854 xylulokinase 232 18 49 1.44 0.2281 4.1.1.37 K01599 uroporphyrinogen decarboxylase 242 90 65 -0.47 0.2292 4.1.99.12 K02858 3,4-dihydroxy 2-butanone 4-phosphate synthase 285 107 77 -0.47 0.2292 2.5.1.49 K01740 O-acetylhomoserine (thiol)-lyase 126 10 27 1.43 0.2304 3.5.2.14 K01474 N-methylhydantoinase B 445 35 94 1.43 0.2304 1.2.1.8 K00130 betaine-aldehyde dehydrogenase 76 6 16 1.42 0.2326 5.3.1.9 K01810 glucose-6-phosphate isomerase 154 12 32 1.42 0.2326 2.6.1.7 cysteine conjugate-beta lyase 2 43 15 11 -0.45 0.2349 3.2.1.31 K01195 beta-glucuronidase 42 15 11 -0.45 0.2349 6.1.1.1 K01866 tyrosyl-tRNA synthetase 397 145 106 -0.45 0.2349 4.4.1.16 selenocysteine lyase 239 19 50 1.4 0.2372 1.8.98.1 K03388 heterodisulfide reductase subunit A 819 296 218 -0.44 0.2377 1.1.1.103 L-threonine 3-dehydrogenase 69 24 18 -0.42 0.2434 1.13.11.52 K00463 indoleamine 2,3-dioxygenase 23 8 6 -0.42 0.2434 6.1.1.7 alanyl-tRNA synthetase 633 225 168 -0.42 0.2434 2.7.7.27 K00975 glucose-1-phosphate adenylyltransferase 287 24 61 1.35 0.2489 3.4.19.1 K01303 acylaminoacyl-peptidase 177 15 38 1.34 0.2512 3.4.21.92 K01358 ATP-dependent Clp protease, protease subunit 361 30 76 1.34 0.2512 2.4.2.29 K00773 queuine tRNA-ribosyltransferase 431 148 113 -0.39 0.252 6.1.1.11 K01875 seryl-tRNA synthetase 386 134 102 -0.39 0.252 2.1.1.72 K06223 DNA adenine methylase 1521 128 322 1.33 0.2536 2.1.1.14 361 122 94 -0.38 0.2549 217 74 57 -0.38 0.2549 163 14 35 1.32 0.256 2.5.1.19 K00549 5-methyltetrahydropteroyltriglutamate-homocysteine methyltransferase K12960 5-methylthioadenosine/S-adenosylhomocysteine deaminase K00057 glycerol-3-phosphate dehydrogenase (NAD(P)+) K00800 3-phosphoshikimate 1-carboxyvinyltransferase 257 22 55 1.32 0.256 2.7.1.5 K00848 rhamnulokinase 72 6 15 1.32 0.256 3.4.16.4 K07260 D-alanyl-D-alanine carboxypeptidase 187 16 40 1.32 0.256 5.3.1.25 K01818 L-fucose isomerase 70 6 15 1.32 0.256 2.7.7.23 290 98 76 -0.37 0.2578 2.7.1.11 K04042 bifunctional UDP-N-acetylglucosamine pyrophosphorylase / Glucosamine-1-phosphate Nacetyltransferase K00850 6-phosphofructokinase 246 21 52 1.31 0.2584 1.2.1.3 K00128 aldehyde dehydrogenase (NAD+) 417 140 109 -0.36 0.2607 2.1.1.80 K00575 chemotaxis protein methyltransferase CheR 723 243 189 -0.36 0.2607 2.3.1.1 K00619 amino-acid N-acetyltransferase 286 95 74 -0.36 0.2607 1.2.1.38 256 22 54 1.3 0.2608 2.7.1.40 K00145 N-acetyl-gamma-glutamyl-phosphate/N-acetylgamma-aminoadipyl-phosphate reductase K00873 pyruvate kinase 303 26 64 1.3 0.2608 2.8.1.1 K01011 thiosulfate/3-mercaptopyruvate sulfurtransferase 174 15 37 1.3 0.2608 6.3.5.4 K01953 asparagine synthase (glutamine-hydrolysing) 368 32 78 1.29 0.2632 4.1.2.13 K01623 fructose-bisphosphate aldolase, class I 563 188 147 -0.35 0.2636 1.4.1.1 alanine dehydrogenase 149 48 38 -0.34 0.2665 4.3.2.1 K01755 argininosuccinate lyase 295 97 77 -0.33 0.2694 3.5.1.25 K01443 N-acetylglucosamine-6-phosphate deacetylase 111 10 24 1.26 0.2705 6.3.5.3 K01952 phosphoribosylformylglycinamidine synthase 595 53 127 1.26 0.2705 3.5.4.1.1.1.94 2.4.1.12 K00694 cellulose synthase (UDP-forming) 89 8 19 1.25 0.2729 4.2.1.113 K02549 O-succinylbenzoate synthase 87 8 19 1.25 0.2729 2.4.2.2 K00756 pyrimidine-nucleoside phosphorylase 66 6 14 1.22 0.2803 6.3.2.12 224 71 58 -0.29 0.2811 6.3.2.17 K11754 dihydrofolate synthase / folylpolyglutamate synthase K01930 folylpolyglutamate synthase 224 71 58 -0.29 0.2811 6.5.1.1 K01971 DNA ligase (ATP) 371 116 95 -0.29 0.2811 1.10.2.2 ubiquinol-cytochrome c reductase hinge protein-like 174 16 37 1.21 0.2828 2.4.2.21 K00768 nicotinate-nucleotide--dimethylbenzimidazole phosphoribosyltransferase K01791 UDP-N-acetylglucosamine 2-epimerase 76 7 16 1.19 0.2878 226 21 48 1.19 0.2878 57 18 15 -0.26 0.2899 3.5.1.81 K00290 saccharopine dehydrogenase (NAD+, L-lysine forming) K06015 N-acyl-D-amino-acid deacylase 57 18 15 -0.26 0.2899 6.3.5.1 NAD synthetase 221 67 56 -0.26 0.2899 1.1.1.81 K00050 hydroxypyruvate reductase 126 12 27 1.17 0.2929 1.17.4.1 K00526 ribonucleoside-diphosphate reductase beta chain 1028 312 262 -0.25 0.2929 2.7.7.8 K00962 polyribonucleotide nucleotidyltransferase 378 36 81 1.17 0.2929 5.3.3.2 K01823 isopentenyl-diphosphate delta-isomerase 167 16 36 1.17 0.2929 6.1.1.17 K01885 glutamyl-tRNA synthetase 348 33 74 1.17 0.2929 6.2.1.3 K01897 long-chain acyl-CoA synthetase 846 256 215 -0.25 0.2929 6.3.5.10 K02232 adenosylcobyric acid synthase 125 12 27 1.17 0.2929 6.1.1.14 K01879 glycyl-tRNA synthetase beta chain 652 62 139 1.16 0.2954 6.1.1.12 K01876 aspartyl-tRNA synthetase 466 139 118 -0.24 0.2959 1.2.1.11 K00133 aspartate-semialdehyde dehydrogenase 298 29 64 1.14 0.3005 2.7.13.3 1679 163 360 1.14 0.3005 5.3.1.23 K07636 two-component system, OmpR family, phosphate regulon sensor histidine kinase PhoR K08963 methylthioribose-1-phosphate isomerase 309 30 66 1.14 0.3005 2.1.2.10 K00605 aminomethyltransferase 266 78 67 -0.22 0.3018 2.6.1.16 K00820 glucosamine--fructose-6-phosphate aminotransferase (isomerizing) 604 178 153 -0.22 0.3018 5.1.3.14 1.5.1.7 3.6.4.6 K06027 vesicle-fusing ATPase 429 125 108 -0.21 0.3048 3.6.3.14 F0F1 ATP synthase subunit epsilon 1767 174 379 1.12 0.3057 1.4.1.13 573 164 144 -0.19 0.3108 1.4.3.4 K00265 glutamate synthase (NADPH/NADH) large chain K00274 monoamine oxidase 70 7 15 1.1 0.3108 6.5.1.4 K01974 RNA 3'-terminal phosphate cyclase 139 14 30 1.1 0.3108 6.2.1.16 K01907 acetoacetyl-CoA synthetase 79 8 17 1.09 0.3134 2.1.2.1 K00600 glycine hydroxymethyltransferase 436 125 110 -0.18 0.3137 6.3.2.9 220 62 55 -0.17 0.3167 2.2.1.7 K01925 UDP-N-acetylmuramoylalanine--D-glutamate ligase 1-deoxy-D-xylulose-5-phosphate synthase 240 67 60 -0.16 0.3197 3.5.2.7 K01468 imidazolonepropionase 109 11 23 1.06 0.3213 6.1.1.20 K01889 phenylalanyl-tRNA synthetase alpha chain 775 216 194 -0.15 0.3228 3.6.3.27 549 57 118 1.05 0.3239 2.6.1.52 K02036 phosphate transport system ATP-binding protein K00831 phosphoserine aminotransferase 80 22 20 -0.14 0.3258 6.3.4.13 K01945 phosphoribosylamine--glycine ligase 324 34 70 1.04 0.3265 1.1.1.38 227 24 49 1.03 0.3292 4.2.3.5 K00027 malate dehydrogenase (oxaloacetatedecarboxylating) K01736 chorismate synthase 297 79 73 -0.11 0.3348 1.1.5.3 sn-glycerol-3-phosphate dehydrogenase subunit C 229 25 50 1 0.3372 2.6.1.82 K09251 putrescine aminotransferase 53 6 12 1 0.3372 3.4.11.2 K01256 aminopeptidase N 165 18 36 1 0.3372 3.6.3.29 158 17 34 1 0.3372 2.7.7.7 K02017 molybdate transport system ATP-binding protein K02338 DNA polymerase III subunit beta 2735 721 675 -0.1 0.3378 3.4.21.66 K08651 thermitase 58 15 14 -0.1 0.3378 3.6.3.16 K01551 arsenite-transporting ATPase 289 76 71 -0.1 0.3378 5.99.1.3 K02470 DNA gyrase subunit B 1690 184 366 0.99 0.3399 2.4.2.11 K00763 nicotinate phosphoribosyltransferase 310 82 77 -0.09 0.3409 2.6.1.9 K00817 histidinol-phosphate aminotransferase 333 87 82 -0.09 0.3409 6.5.1.2 K01972 DNA ligase (NAD+) 290 32 63 0.98 0.3426 1.6.5.3 K00330 NADH dehydrogenase I subunit A 4484 1170 1104 -0.08 0.3439 6.1.1.19 arginyl-tRNA synthetase 444 115 109 -0.08 0.3439 3.5.1.18 K01439 succinyl-diaminopimelate desuccinylase 253 28 55 0.97 0.3453 3.5.4.26 K11752 diaminohydroxyphosphoribosylaminopyrimidine deaminase / 5-amino-6-(5-phosphoribosylamino)uracil reductase K13953 alcohol dehydrogenase, propanol-preferring 165 43 41 -0.07 0.3469 743 83 161 0.96 0.348 170 19 37 0.96 0.348 4.4.1.11 K00590 site-specific DNA-methyltransferase (cytosineN4-specific) K01761 methionine-gamma-lyase 91 23 22 -0.06 0.3499 4.3.1.19 K01754 threonine dehydratase 238 27 52 0.95 0.3507 2.3.2.2 K00681 gamma-glutamyltranspeptidase 230 26 50 0.94 0.3534 1.11.1.5 K00428 cytochrome c peroxidase 95 11 21 0.93 0.3561 2.6.1.19 274 32 60 0.91 0.3616 1.1.1.23 K07250 4-aminobutyrate aminotransferase / (S)-3amino-2-methylpropionate transaminase K00013 histidinol dehydrogenase 295 73 72 -0.02 0.3621 3.6.4.13 K05592 ATP-dependent RNA helicase DeaD 278 33 61 0.89 0.3671 4.1.2.9 K01621 phosphoketolase 59 7 13 0.89 0.3671 6.3.1.2 K01915 glutamine synthetase 558 66 122 0.89 0.3671 3.4.11.1 K01255 leucyl aminopeptidase 241 59 59 0 0.3682 3.6.4.3 K13254 spastin 79 19 19 0 0.3682 4.1.3.39 K01666 4-hydroxy 2-oxovalerate aldolase 25 6 6 0 0.3682 1.1.1.14 K00008 L-iditol 2-dehydrogenase 141 17 31 0.87 0.3727 1.3.99.1 succinate dehydrogenase flavoprotein subunit 752 90 164 0.87 0.3727 4.2.1.120 4-hydroxyphenylacetate-3-hydroxylase (HpaA-1) 50 6 11 0.87 0.3727 4.99.1.3 K03795 sirohydrochlorin cobaltochelatase 48 6 11 0.87 0.3727 4.1.1.31 K01595 phosphoenolpyruvate carboxylase 292 70 71 0.02 0.3743 5.1.1.1 K01775 alanine racemase 261 62 63 0.02 0.3743 1.1.1.1 2.1.1.113 1.4.4.2 K00282 glycine dehydrogenase subunit 1 620 147 150 0.03 0.3774 2.5.1.15 K00796 dihydropteroate synthase 244 30 54 0.85 0.3783 4.2.1.49 K01712 urocanate hydratase 167 39 40 0.04 0.3804 3.2.1.4 K01179 endoglucanase 320 39 70 0.84 0.3811 4.1.99.1 K01667 tryptophanase 75 9 16 0.83 0.3839 1.1.1.18 K00010 myo-inositol 2-dehydrogenase 110 25 26 0.06 0.3865 3.4.23.42 K01385 thermopsin 95 22 23 0.06 0.3865 1.4.1.3 K00261 glutamate dehydrogenase (NAD(P)+) 336 42 74 0.82 0.3867 2.7.9.1 K01006 pyruvate,orthophosphate dikinase 403 50 88 0.82 0.3867 5.4.2.8 K00966 mannose-1-phosphate guanylyltransferase 352 44 77 0.81 0.3895 6.1.1.22 asparaginyl-tRNA synthetase 284 36 63 0.81 0.3895 6.1.1.6 lysyl-tRNA synthetase 488 61 107 0.81 0.3895 6.3.5.2 GMP synthase 519 65 114 0.81 0.3895 6.3.4.4 K01939 adenylosuccinate synthase 336 77 81 0.07 0.3896 5.4.99.2 K01849 methylmalonyl-CoA mutase, C-terminal domain 556 70 122 0.8 0.3923 5.4.2.1 K01834 phosphoglycerate mutase 719 163 172 0.08 0.3926 1.1.1.261 K00096 glycerol-1-phosphate dehydrogenase [NAD(P)] 117 15 26 0.79 0.3951 2.3.1.179 K09458 3-oxoacyl-[acyl-carrier-protein] synthase II 311 40 69 0.79 0.3951 1.2.1.70 K02492 glutamyl-tRNA reductase 195 43 46 0.1 0.3987 6.3.5.9 K02224 cobyrinic acid a,c-diamide synthase 132 29 31 0.1 0.3987 1.8.1.4 K00382 dihydrolipoamide dehydrogenase 480 62 106 0.77 0.4008 3.4.17.19 K01299 carboxypeptidase Taq 206 27 46 0.77 0.4008 4.1.2.27 K01634 sphinganine-1-phosphate aldolase 53 12 13 0.12 0.4049 1.2.1.16 167 22 37 0.75 0.4065 2.4.1.21 K00135 succinate-semialdehyde dehydrogenase (NADP+) K00703 starch synthase 339 73 80 0.13 0.4079 3.4.11.14 peptidase T 52 11 12 0.13 0.4079 1.14.99.30 zeta carotene desaturase candidate 45 6 10 0.74 0.4094 2.3.1.29 K00639 glycine C-acetyltransferase 135 18 30 0.74 0.4094 3.4.11.4 peptidase T 44 6 10 0.74 0.4094 6.1.1.3 K01868 threonyl-tRNA synthetase 498 66 110 0.74 0.4094 6.2.1.26 K01911 O-succinylbenzoic acid--CoA ligase 66 9 15 0.74 0.4094 3.6.4.12 K03655 ATP-dependent DNA helicase RecG 2293 493 544 0.14 0.411 3.5.3.12 K10536 agmatine deiminase 43 9 10 0.15 0.414 2.4.1.25 K00705 4-alpha-glucanotransferase 280 38 62 0.71 0.418 1.2.1.2 K00123 formate dehydrogenase, alpha subunit 660 139 156 0.17 0.4202 2.2.1.6 K01652 acetolactate synthase I/II/III large subunit 610 128 144 0.17 0.4202 2.6.1.87 uridine 5'-(beta-1-threo-pentapyranosyl-4-ulose diphosphate) aminotransferase, PLP-dependent K01668 tyrosine phenol-lyase 230 49 55 0.17 0.4202 40 8 9 0.17 0.4202 211 29 47 0.7 0.4209 2.3.1.9 K00974 tRNA nucleotidyltransferase (CCA-adding enzyme) K00626 acetyl-CoA C-acetyltransferase 773 161 182 0.18 0.4232 2.7.7.6 K03040 DNA-directed RNA polymerase subunit alpha 2627 361 584 0.69 0.4238 3.1.4.3 K01114 phospholipase C 32 7 8 0.19 0.4263 4.3.1.1 K01744 aspartate ammonia-lyase 69 14 16 0.19 0.4263 1.3.1.34 K13237 peroxisomal 2,4-dienoyl-CoA reductase 74 10 16 0.68 0.4267 2.1.1.79 K00574 cyclopropane-fatty-acyl-phospholipid synthase 70 10 16 0.68 0.4267 4.2.1.20 K01696 tryptophan synthase beta chain 718 100 160 0.68 0.4267 1.1.1.35 K07516 3-hydroxyacyl-CoA dehydrogenase 240 34 54 0.67 0.4296 2.7.1.30 K00864 glycerol kinase 159 22 35 0.67 0.4296 6.3.3.1 235 33 52 0.66 0.4325 3.2.1.3 K01933 phosphoribosylformylglycinamidine cycloligase K01178 glucoamylase 97 14 22 0.65 0.4354 4.1.2.5 K01620 threonine aldolase 98 14 22 0.65 0.4354 4.2.1.79 2-methylcitrate dehydratase 100 14 22 0.65 0.4354 6.1.1.5 K01870 isoleucyl-tRNA synthetase 763 108 170 0.65 0.4354 2.3.1.16 K07513 acetyl-CoA acyltransferase 1 183 37 43 0.22 0.4355 4.1.99.2 2.7.7.72 2.4.1.129 K05364 peptidoglycan glycosyltransferase 657 132 154 0.22 0.4355 5.4.2.7 K01839 phosphopentomutase 61 9 14 0.64 0.4383 2.4.1.1 K00688 starch phosphorylase 396 57 88 0.63 0.4413 5.4.99.16 K05343 maltose alpha-D-glucosyltransferase 56 11 13 0.24 0.4416 6.1.1.9 K01873 valyl-tRNA synthetase 690 136 161 0.24 0.4416 3.4.24.57 K01409 O-sialoglycoprotein endopeptidase 333 48 74 0.62 0.4442 4.2.3.4 K01735 3-dehydroquinate synthase 204 30 46 0.62 0.4442 6.3.5.7 1149 167 257 0.62 0.4442 1.3.99.2 K03330 glutamyl-tRNA(Gln) amidotransferase subunit E K00248 butyryl-CoA dehydrogenase 401 79 94 0.25 0.4446 5.5.1.4 K01858 myo-inositol-1-phosphate synthase 269 53 63 0.25 0.4446 1.3.99.3 K00249 acyl-CoA dehydrogenase 546 106 127 0.26 0.4477 5.99.1.2 K03168 DNA topoisomerase I 568 111 133 0.26 0.4477 1.1.1.157 K00074 3-hydroxybutyryl-CoA dehydrogenase 252 37 56 0.6 0.4501 2.7.7.19 K00970 poly(A) polymerase 304 45 68 0.6 0.4501 3.6.1.3 K01509 adenosinetriphosphatase 673 129 157 0.28 0.4538 2.6.1.44 108 16 24 0.58 0.456 3.4.13.9 K00827 alanine-glyoxylate transaminase / (R)-3-amino2-methylpropionate-pyruvate transaminase K01271 Xaa-Pro dipeptidase 199 30 45 0.58 0.456 1.17.7.1 4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase 203 31 46 0.57 0.4589 1.13.11.5 K00451 homogentisate 1,2-dioxygenase 64 12 15 0.32 0.466 1.2.7.1 ferredoxin oxidoreductase alpha subunit 1069 199 248 0.32 0.466 2.3.1.15 K13508 glycerol-3-phosphate acyltransferase 260 48 60 0.32 0.466 2.3.3.1 K01647 citrate synthase 394 73 91 0.32 0.466 5.5.1.1 K01856 muconate cycloisomerase 42 8 10 0.32 0.466 4.2.1.2 K01679 fumarate hydratase, class II 458 71 103 0.54 0.4678 6.1.1.4 K01869 leucyl-tRNA synthetase 793 123 179 0.54 0.4678 1.1.1.193 K00082 5-amino-6-(5-phosphoribosylamino)uracil reductase K00058 D-3-phosphoglycerate dehydrogenase 235 43 54 0.33 0.469 632 116 146 0.33 0.469 1.1.1.95 1.1.1.85 K00052 3-isopropylmalate dehydrogenase 262 41 59 0.53 0.4708 2.7.7.63 K03800 lipoate-protein ligase A 269 49 62 0.34 0.472 1.1.1.42 K00031 isocitrate dehydrogenase 365 58 83 0.52 0.4738 1.5.3.1 N-methyltryptophan oxidase 191 30 43 0.52 0.4738 1.14.13.81 120 19 27 0.51 0.4767 3.4.21.102 K04035 magnesium-protoporphyrin IX monomethyl ester (oxidative) cyclase K03797 carboxyl-terminal processing protease 341 54 77 0.51 0.4767 2.7.2.3 phosphoglycerate kinase 372 67 86 0.36 0.4781 5.2.1.8 K03768 peptidyl-prolyl cis-trans isomerase B (cyclophilin B) CTP synthetase 445 80 103 0.36 0.4781 514 83 117 0.5 0.4797 173 31 40 0.37 0.4811 95 15 21 0.49 0.4827 871 141 198 0.49 0.4827 4.2.1.39 K01929 UDP-N-acetylmuramoylalanyl-D-glutamyl-2,6diaminopimelate--D-alanyl-D-alanine ligase K00021 hydroxymethylglutaryl-CoA reductase (NADPH) K02434 aspartyl-tRNA(Asn)/glutamyl-tRNA (Gln) amidotransferase subunit B hypothetical protein 59 10 13 0.38 0.4842 5.4.2.2 K01835 phosphoglucomutase 181 32 42 0.39 0.4872 1.1.1.205 K00088 IMP dehydrogenase 387 64 88 0.46 0.4917 1.1.1.47 K00034 glucose 1-dehydrogenase 53 9 12 0.42 0.4963 1.6.1.2 K00324 NAD(P) transhydrogenase subunit alpha 158 27 36 0.42 0.4963 6.3.4.2 6.3.2.10 1.1.1.34 6.3.5.6