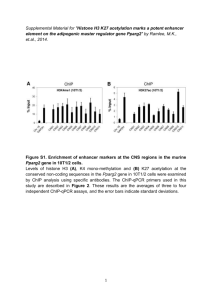
Modern definition of epigenetics is as follows: it is non
advertisement
Molecules and mechanisms of epigenetics Basis of epigenetics Adult stem cells know their fate! For example: myoblasts can form muscle cells only. Hematopoetic cells only become blood cells. .....but all those cells have identical genome sequences! Modern definition of epigenetics is as follows: it is nonsequence dependent inheritance and chromatine changes. How can just paternal or maternal traits be expressed in offspring? This is called genetic imprinting. How can females express only one X chromosome per cell? We observe some changes in gene expression that are heritable! Inactive X has unacetylated histone H4 Epigenetics is based on four different mechanisms (so far…..) 1. DNA Methylation 2. Chromatin remodelling 3. Histone modification 4. RNA interference/interactions Two faces of chromatin control DNA from zygote to adult – methylation state Chromatin restricts accessibility of the genome Horn and Peterson, Science, 2002 Euchromatin Transcriptionally active, less compacted Heterochromatin Less transcriptionally active, very compacted a) constitutive heterochromatin centromeres, telomeres b) facultative heterochromatin rDNA, transposons, inactive X chromosome Condensed chromatin state depends on cytosine methylation Daug Brutlag, 2011 Two most obvious states of chromatin Importance of CpG Sequences in CpG Islands Near Promoters Architecture of Epigenome DNA is compacted through interactions with proteins DNA and chromatine proteins is a template for epigenetic phenomena Numerous post-translational histone modifications Histone acetylation Acetylation of the lysine residues at the N terminus of histons removes positive charges, thereby reducing the affinity between histones and DNA. This makes RNA polymerase and transcription factors easier to access the promoter region. Therefore, in most cases, histone acetylation enhances transcription while histone deacetylation represses transcription. Histone acetylation is catalyzed by histone acetyltransferases (HATs) and histone deacetylation is catalyzed by histone deacetylases (denoted by HDs or HDACs). Several different forms of HATs and HDs have been identified. Among them, CBP/p300 is probably the most important, since it can interact with numerous transcription regulators. Importance of acetylation at the histon level Acetylation of histones serves as epigenetic marker within chromatin. Several reports have shown that one modification has the tendency to influence whether another modification will take place. Modifications of histones can not only cause secondary structural changes at their specific points, but can cause many structural changes in distant locations which inevitably affects function. As the chromosome is replicated, the modifications that exist on the parental chromosomes are handed down to daughter chromosomes. The modifications, as part of their function, can recruit enzymes for their particular function and can contribute to the continuation of modifications and their effects after replication has taken place. It has been shown that, even past one replication, expression of genes may still be affected many cell generations later. Histone Methylation Unlike acetylation, histone methylation does not alter the charge of the modified residues and it is therefore less likely to directly alter nucleosomal interactions required for chromatin folding. This probably explains why histone methylation can either repress or activate transcription depending on location. Arginine methylation of histone H3 and H4 promotes transcriptional activation, whereas lysine methylation of histone H3 and H4 is implicated in both transcriptional activation and repression, depending on the methylation site. In addition, lysine residues can be methylated in the form of mono-, di-, or tri-methylation, providing further functional diversity to each site of lysine methylation. For example, trimethylation on K4 of Histone H3 (H3K4me3) is generally associated with transcriptional activation, whereas tri-methylation on K9 and K27 of histone H3 (H3K9me3 & H3K27me3) are generally associated with transcriptional repression. For many years, histone methylation was thought to be irreversible as it is a stable mark propagated through multiple cell divisions. However, it was recently shown that, similarly to histone acetylation, methylation is an actively regulated and reversible process. Histone code hypothesis The Histone code hypothesis suggests the idea that patterns of post-translational modifications on histones, collectively, can direct specific cellular functions. Chemical modifications of histone proteins often occur on particular amino acids. This specific addition of single or multiple modifications on histone cores can be interpreted by transcription factors and complexes which leads to functional implications. This process is facilitated by enzymes such as HATs and HDACs that add or remove modifications on histones, and transcription factors that process and "read" the modification codes. The outcome can be activation of transcription or repression of a gene. For example, the combination of acetylation and phosphorylation have synergistic effects on the chromosomes overall structural condensation level and, hence, induces transcription activation of immediate early gene. Experiments investigating acetylation patterns of H4 histones suggested that these modification patterns are collectively maintained in mitosis and meiosis in order to modify long-term gene expression. The acetylation pattern is regulated by HAT and HADC enzymes and, in turn, sets the local chromatin structure. In this way, acetylation patterns are transmitted and interconnected with protein binding ability and functions in subsequent cell generation.