Ch 5: Protein Synthesis
advertisement
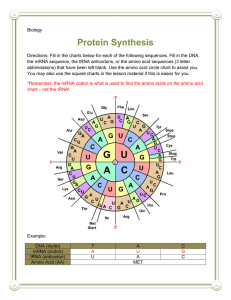
Protein Synthesis 3.5, 7.3, 7.4 Remember Gregor Mendel? Mendel’s experiments with garden peas led to the foundations of genetics Hypothesized that characteristics such as height, shape and colour of the seed, colour of flowers, etc were inherited factors. Today we call those factors GENES (A gene is a sequence of nucleotides in DNA that performs a specific function.) Genes direct production of PROTEINS Proteins: - antibodies - hormones - enzymes that drive cellular processes - determine physical characteristics - manifest genetic disorders Nt -> amino acids -> proteins One gene- one polypeptide A single gene codes for one polypeptide (protein) Some proteins consist of more than one polypeptide chain (ex: hemoglobin) Each chain is controlled by a different gene. p235 P 236 Overview of Protein Synthesis DNA contains the blueprints for making proteins. The sequence of nucleotides must be read by a ribosome to determine the sequence of amino acids to make the protein. DNA is found in the nucleus but protein synthesis takes place in the cytoplasm Problems: DNA can’t leave the nucleus Could be damaged if it did which would lead to the death of the cell (possibly the organism) Inefficient if DNA was read by ribosomes because then only 1 protein from a single strand could be created at a time. A mechanism has evolved for protein synthesis that prevents DNA from having to leave the cell. TRANSCRIPTION TRANSLATION nucleus OVERVIEW 1. TRANSCRIPTION – DNA is transcribed into mRNA in the nucleus 2. mRNA exits the nucleus and enters the cytoplasm 3. TRANSLATION – mRNA is “read” by ribosomes to make the polypeptide chain 4. Polypeptide chain is released and folds into a protein. mRNA strand can be used again. DNA vs RNA Deoxyribonucleic acid Pentose sugar: deoxyribose (1 less O) Double stranded N bases: A,C,G, T (no uracil!) A–T In nucleus Ribonucleic acid Pentose sugar: ribose Single stranded N bases: A,C,G and URACIL (U) (no thymine!) A–U In nucleus and in cytoplasm RNA 1) 2) 3) There are 3 types of RNA mRNA: messenger RNA – an RNA version of a gene tRNA: transfer RNA – brings the appropriate amino acid to the ribosome rRNA: ribosomal RNA – structural component of ribosomes (involved in assembly of polypeptide) The Genetic Code The building blocks of proteins are amino acids. There are 20 amino acids A sequence of 3 nucleotides called a CODON code for different amino acids. There are 64 different codons (43 = 64) Therefore, there may be more than one codon for a given amino acid. Ser Ser Ser Ser p43 The first codon is the “start codon” It signals the start of protein synthesis. In almost all polypeptides, AUG is the start codon. AUG also codes for methionine. (GUG and UUG are sometimes act as start codons) There are 3 “stop codons”: UAA, UAG, UGA These do not code for amino acids but stop synthesis. Homework: Read 5.1 and 5.2 5.1 p236: Q: 5 5.2 page 241: Q’s: 1,3-5,8,10-13 Transcription Transcription The process of making an mRNA strand from the DNA gene sequence 3 steps: initiation, elongation, termination INITIATION The enzyme RNA polymerase attaches to the DNA double helix at the promoter region. PROMOTER REGION: A sequence of DNA “upstream” from the gene that contains a lot of A-T base pairs. (easier to break apart than G-C) Initiation con’t RNA polymerase unwinds and separates the helix exposing the template strand. Since RNA polymerase only binds at the promoter region, it allows only the gene to be transcribed. Rich in A-T Coding strand ELONGATION Once the helix is open, RNA polymerase starts building a single stranded mRNA in the 5’-3’ direction using RNA nucleotides and complimentary base pairing rules. Only one strand of DNA is used. This is the template strand. Elongation con’t…. The other DNA strand is the coding strand This strand is identical to the mRNA strand except that it has thymine instead of uracil. Note: the promoter region does not get transcribed. As the DNA is transcribed, it is rewound into its helix Termination The mRNA strand is synthesized until RNA polymerase reaches the terminator sequence at the end of the gene. The mRNA strand will detach from the DNA template. This mRNA strand is called the primary transcript Post-transcription After transcription, the mRNA primary transcript must go to the cytoplasm to be translated. In eukaryotes, before it leaves the nucleus it needs a “cap” and a “tail” to protect it and prevent it from being broken down. 5’ CAP A 5’ cap is added to mRNA. This is a modified guanine nucleoside triphosphate. The cap protects the mRNA from digestion from enzymes when it enters the cytoplasm Poly-A tail A sequence of ~ 200 adenine ribonucleosides is added to the 3’ end to protect the mRNA from breaking down This is called a poly-A tail The tail is added with the help of the enzyme poly-A polymerase Exons and Introns In eukaryotic DNA, a gene consists of introns and exons EXONS: coding regions – they code for the specific protein INTRONS: noncoding – “filler DNA” that does not code for proteins. Introns and Exons con’t… Before mRNA goes into the cytoplasm, the introns must be removed from the mRNA Proteins called spliceosomes cut the introns out and join the remaining exons together. mRNA transcript Once the cap and tail have been added, and the introns removed, the mRNA is called an mRNA transcript It can now go to the cytoplasm for translation Unlike in DNA replication, there is no enzyme to “spell check” the mRNA. But errors are not too detrimental since multiple mRNA transcripts are made for each gene so sufficient amounts of protein will be produced by the correct copies. TRANSLATION Remember….. Protein synthesis = the making of a protein chain (polypeptide) A protein chain is made of several amino acids bonded together. The order of the amino acids in a protein is important to the overall shape of the protein. Therefore, it is important that the correct amino acids be placed in the correct sequence. Step # 1: Transcription: a template strand of DNA is used to create an mRNA strand Template strand (antisense strand) 3’ CCT TAG GTA GGT ATT 5’ 5’ GGA ATG CAT CCA TAA 3’ Coding strand (sense strand) 5’ GGA AUG CAU CCA UAA 3’ mRNA transcript Remember, the primary mRNA transcript will have introns that need to be removed. Also a cap and a tail will be added before the mRNA can leave the nucleus. Step #2: Translation: Following the sequence determine by the mRNA strand, tRNA brings amino acids to the ribosome in the correct order to create the polypeptide. How does an amino acid bond to the tRNA? tRNA (transfer RNA) – a clover shaped molecule made of RNA nucleotides. This 3’ end is the acceptor stem. This is where the amino acid will bind. Only one type of amino acid will bind to any given tRNA. This is the anticodon. The anticodon is the codon that determines which amino acid will bind to the tRNA strand at the acceptor stem. The anticodon is complimentary to the codon on the mRNA strand that codes for the amino acid. Ex: the codon for methionine is AUG, so if this tRNA wanted to carry methionine, its anticodon would be UAC. For the amino acid to bind at the acceptor stem, the process is catalyzed by a tRNAactivating enzyme. There are 20 different amino acids, and so 20 different tRNA activating enzymes. The amino acid reacts with the adenine of the CCA codon at the acceptor stem to form a covalent bond through a condensation reaction. How many different tRNA molecules are there? How many different tRNA molecules are there? There are 64 different codons, however, 3 of those codons are stop codons and do not code for an amino acid, and so tRNAs do not exist for these. Therefore, there are 61 different tRNA molecules. How many tRNA activating enzymes are there? 1 for every amino acid, so 20 What are ribosomes? Ribosomes are the organelle where translation takes place. They are extremely small, but abundant. Ribosomes hold the mRNA strand in place and are where the tRNA bind to the mRNA, bringing with it the appropriate amino acid. Ribsosomes are made or ribosomal RNA (rRNA) and proteins Ribosomes Ribosomes consist of 2 subunits: The “small subunit” is made of 1 rRNA molecules (and proteins) This is where mRNA binds The “large subunit”: is made of 2 rRNA molecules This is where the tRNA binds Has the A, P, and E sites When the ribosomes are not translating, the subunits are separated. They only come together for translation. Occasionally, several ribosomes may be attached to a single mRNA strand, with each ribosome constructing a polypeptide chain. In This way, several polypeptides can be made simultaneously. A, P, E sites of Ribosome A = “attachment” site: this is where tRNA enters the ribosome and attaches to the mRNA strand P = “peptidyl” site: where peptide bonds formed E = “exit” site: where the tRNA detaches from the ribosome and the mRNA strand Translation Occurs in the 5’ – 3’ direction of mRNA Divided into 3 stages: initiation, elongation , termination INITIATION the ribosome attaches to mRNA The start codon on the mRNA is AUG. So the 1sttRNA required will have the anticodon UAC and the amino acid methionine attached to it. Initiation The tRNA will bind to the base of the P site on the small subunit of the ribosome. This is catalyzed with the help of the enzyme ATPase. The rRNA of the small subunit will attach to a binding site on the mRNA. Initiation The small subunit slides along the mRNA until the P site reaches the start codon (AUG). Then the larger subunit, containing the binding sites for the tRNA, join the complex. ELONGATION building of the protein strand with the 1sttRNA (carrying met) in the P site, the next tRNA can bind to the A site. (All tRNA bind to the ribosome with the help of ATPase) Elongation The amino acid methionine of the 1st tRNA will detach from the tRNA and form a peptide bond with the amino acid attached to the 2nd tRNA which is in the A site. This is catalyzed by the enzyme peptidyltransferase. Elongation The ribosome moves down the mRNA chain toward the 3’end. Now the 1st tRNA no longer has its amino acid and it is now in the E site and can be released from the ribosome. TERMINATION release from the ribosome complex The stop codon will be near the 3’ end of the mRNA strand When a stop codon is in the A site, there is no tRNA available and this is a signal to end protein synthesis. Termination A “protein release factor” will come to release the last tRNA from the last amino acid. The polypeptide can now be released from the ribosome. The ribosome will disassociate into the small and large subunits again and can be used again and again. The tRNAs and the mRNA can also be used again. Animations http://highered.mcgrawhill.com/sites/0072437316/student_view0/ chapter15/animations.html# http://www.youtube.com/watch?v=NJxobg kPEAo&feature=email http://www.youtube.com/watch?v=B6O6u Rb1D38&list=PL1AD35ADA1E93EB6F Ribosome Distribution If the protein being synthesized is to be used inside the cell by the cell, the mRNA will use ribosomes scattered throughout the cytoplasm. If the protein being synthesized is to be exported out of the cell or used by lysosomes, the mRNA will use ribosomes attached to the endoplasmic reticulum. They will then go to a Golgi complex to be packaged into vesicles (have a membrane place around it).